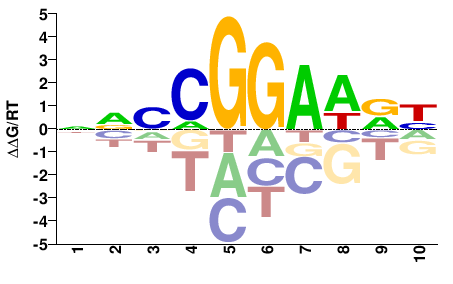
# Parameters
data_name = "Elk4"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:04:03,911 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Elk4: #244
| Cell type | Modisco | Homer |
|---|---|---|
| MPP;A;CL |
Significance: 0.030398599999999998 |
Significance: 0.00688714 |
| naiveCD8;A;CL |
Significance: 0.00251489 |
Significance: 0.00212991 |
| memT;CD4+;A;CL |
Significance: 0.00163719 |
Significance: 8.895169999999999e-06 |
| Th17;CD4+;A;CL |
Significance: 0.00217904 |
Significance: 0.0013694 |
| NK_SPLND;A;CL |
Significance: 0.00132184 |
Significance: 0.000525425 |
| preB;A;CL |
Significance: 0.000454707 |
Significance: 0.000387756 |
| immatureB;A;CL |
Significance: 0.00185359 |
Significance: 0.000325531 |
| matureB;A;CL |
Significance: 0.0013090999999999999 |
Significance: 0.000493315 |
| MZ_B;A;CL |
Significance: 0.013435599999999999 |
Significance: 0.00285211 |
| B1_B;A;CL |
Significance: 0.00286579 |
Significance: 0.000180407 |
| GC_B;A;CL |
Significance: 0.0539359 |
Significance: 0.000867457 |
| plasmaB;A;CL |
Significance: 0.03561690000000001 |
Significance: 0.000247057 |
| aB_LPS;A;CL |
Significance: 0.000676143 |
Significance: 0.000365574 |
| aB_LPS+IL4;A;CL |
Significance: 0.00102051 |
Significance: 0.00041282199999999997 |
| aB_LPS+IL21;A;CL |
Significance: 0.00693232 |
Significance: 0.00015493 |
| aB_LSP+a_d_dextran;A;CL |
Significance: 0.00036221699999999996 |
Significance: 0.0010831 |
| aB_CD40+IL4;A;CL |
Significance: 0.00015488 |
Significance: 0.00034561199999999997 |
| Mast;A;CL |
Significance: 0.00150522 |
Significance: 0.00101618 |
| BSPHL;A;CL |
Significance: 0.00333477 |
Significance: 0.00137747 |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 0.00019553900000000003 |
Significance: 0.000917506 |
| ESNPHL_SPLND_BlbCA;CL |
Significance: 0.00468635 |
Significance: 0.00012035 |
| NTRPHL;A;CL |
Significance: 0.00587973 |
Significance: 0.000521135 |
| Tfh_LCMV;A;CL |
Significance: 0.00416526 |
Significance: 5.74355e-06 |
| Tfh;A;CL |
Significance: 0.00266671 |
Significance: 0.00034050300000000003 |
| Tfh_mut;A;CL |
Significance: 0.00394488 |
Significance: 0.000449116 |
| aNonTfh;A;CL |
Significance: 0.000900447 |
Significance: 3.49114e-06 |
| aNonTfh_mut;A;CL |
Significance: 0.000992723 |
Significance: 8.85487e-07 |
| Tfr;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.00052244 |
Significance: 5.9571300000000005e-06 |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.000600763 |
Significance: 1.9354300000000002e-05 |
| naiveT;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.00105533 |
Significance: 0.000125654 |
| naiveT_mut;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.00124758 |
Significance: 4.12183e-07 |
| largeDPprecrsr;CD4+CD8+CD69-;A;CL |
Significance: 0.00120143 |
Significance: 0.00081162 |
| smallDPprecrsor;CD4+CD8+CD69-;A;CL |
Significance: 0.00291213 |
Significance: 1.12048e-05 |
| CD4+CD8intTCR+CD69+CD24+;B2defM;A;CL |
Significance: 0.00388392 |
Significance: 0.000105531 |
| CD4+CD8-TCR+CD69+CD24+CD25-;B2defM;A;CL |
Significance: 0.001144 |
Significance: 5.10651e-05 |
| CD4+CD8-TCR+CD69-CD24-CD25-;B2defM;A;CL |
Significance: 0.00107283 |
Significance: 5.70936e-07 |
| CD4+CD8intTCR+CD69+CD24+;MHC2defM;A;CL |
Significance: 0.00203587 |
Significance: 3.0164299999999996e-06 |
| CD4+CD8+TCR+CD69+CD24+;MHC2defM;A;C |
Significance: 0.00514752 |
Significance: 0.00120291 |
| CD4+CD8+TCR+CD69-CD24-;MHC2defM;A;CL |
Significance: 0.00165594 |
Significance: 0.00208086 |
| CD4+CD8-TCR+CD25+Foxp3-;A;CL |
Significance: 0.00229605 |
Significance: 3.63e-06 |
| CD4+CD8-TCR+CD25-Foxp3+;A;CL |
Significance: 0.00149493 |
Significance: 8.81695e-08 |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL |
Significance: 0.00280439 |
Significance: 3.71981e-05 |
| DC_BMD;A;CL |
Significance: 0.0007455989999999999 |
Significance: 0.038348 |
| MCRPHG;A;CL |
Significance: 0.00437295 |
Significance: 0.0344824 |
| Erythroblast;A;CL |
Significance: 0.00188017 |
Significance: 0.00317101 |
| DP;A;CL |
Significance: 0.00281668 |
Significance: 2.70705e-07 |
| DP3a;A;CL |
Significance: 0.00922575 |
Significance: 0.00352946 |
| DP3b;A;CL |
Significance: 0.00618203 |
Significance: 0.000454275 |
| ISP;A;CL |
Significance: 0.00146092 |
Significance: 0.00915257 |
| ES;MEF;A;CL |
Significance: 0.00129049 |
Significance: 0.00268033 |
| SML_INTSTN;A;CL |
Significance: 0.000436727 |
Significance: 0.000413605 |
| LRG_INTSTN;A;CL |
Significance: 0.00179064 |
Significance: 0.00280316 |
| LNG;A;CL |
Significance: 7.243930000000001e-05 |
Significance: 0.000311565 |
| Muscle;A;CL |
Significance: 0.000109006 |
Significance: 0.029725599999999998 |
| Heart;D;En |
Significance: 0.000556474 |
Significance: 0.00440778 |
| Adipose;A;CL |
Significance: 0.00826674 |
Significance: 0.00028167 |
| Mammary;A;CL |
Significance: 0.00230133 |
Significance: 0.00109259 |
| Adrenal;A;CL |
Significance: 0.000117837 |
Significance: 0.0451026 |
| testis;A;CL |
Significance: 0.00277298 |
Significance: 0.0015566 |
| placenta;A;CL |
Significance: 0.00111082 |
Significance: 0.00233988 |
| SPNL_CHRD;A;CL |
Significance: 0.000474696 |
Significance: 0.00326983 |
| STMCH;A;CL |
Significance: 0.000189564 |
Significance: 0.00281195 |
| PNCRS;A;CL |
Significance: 0.0006100319999999999 |
Significance: 0.0007978560000000001 |
| eye-NRL_RTN;A;CL |
Significance: 0.000313271 |
Significance: 0.018448 |
| NRV;Eye-OPTC;A;CL |
Significance: 3.38209e-05 |
Significance: 0.0116596 |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.0435935 |
Significance: 0.0443881 |
| BRN;FRNTL_CRTX;A;CL |
Significance: 8.457540000000001e-05 |
Significance: 0.057783600000000004 |
| BRN;PTMN;A;CL |
Significance: 0.0269983 |
Significance: 0.0237793 |
| BRN;MDL_OBLNGT;A;CL |
Significance: 0.00043678099999999996 |
Significance: 0.0108363 |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.000156315 |
Significance: 0.0245183 |
| BRN;CRBLM;A;CL |
Significance: 0.000422674 |
Significance: 0.00517718 |
| restingB;A;CL |
Significance: 0.000137048 |
Significance: 1.5240899999999998e-05 |
| aB;CD180;A;CL |
Significance: 0.00068904 |
Significance: 4.3987200000000005e-05 |
| LNG_TRM_HSC;A;CL |
Significance: 0.00163347 |
Significance: 5.94404e-05 |
| SHRT_TRM_HSC;A;CL |
Significance: 0.0227051 |
Significance: 0.00245254 |
| gamma_delta_TCR;A;CL |
Significance: 0.00019898799999999999 |
Significance: 5.20856e-05 |
| gamma_delta_TCR_MCR;A;CL |
Significance: 0.00470721 |
Significance: 0.000632216 |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.00107615 |
Significance: 6.76845e-07 |
| NK;IL2+IL12+;A;CL |
Significance: 0.00180464 |
Significance: 0.0023497 |
| ILC2precrsr;A;CL |
Significance: 0.00129937 |
Significance: 0.00297519 |
| NCR+ILC3;A;CL |
Significance: 0.0033779000000000005 |
Significance: 0.0005568530000000001 |
| ILC2_Nippo;A;CL |
Significance: 0.00131082 |
Significance: 0.00037517599999999996 |
| ILC1;A;CL |
Significance: 0.00301379 |
Significance: 0.00176594 |
| NCR+ILC3+IL23;A;CL |
Significance: 0.00424871 |
Significance: 0.0009102 |
| ILC2;A;CL |
Significance: 0.00173396 |
Significance: 1.5149299999999999e-06 |
| Th2_nippo;A;CL |
Significance: 0.00136197 |
Significance: 0.00033502900000000003 |
| aB_LPS+IL4_24h;A;CL |
Significance: 7.21609e-05 |
Significance: 0.00127643 |
| aB_LPS+IL4+Oligomycin_24h;A;CL |
Significance: 0.0328517 |
Significance: 0.00116928 |
| CH12_QNTPLE;A;CL |
Significance: 0.000147955 |
Significance: 0.000480939 |
| wCH12;A;CL |
Significance: 0.05307919999999999 |
Significance: 0.000601079 |
| CH12;med14;A;CL |
Significance: 0.00124349 |
Significance: 0.00038041 |
| wCH12_drug;A;CL |
Significance: 0.000260447 |
Significance: 0.000535379 |
| CLP;A;CL |
Significance: 0.00801821 |
Significance: 0.025799700000000002 |
| mem_precrsrT_SPLND;CD8+;A;GEO |
Significance: 0.00138119 |
Significance: 0.00155999 |
| NK_LVRD;CD19-CD3e-NKp46+DX+Trail-;A;GEO |
Significance: 0.00257044 |
Significance: 0.00667934 |
| NTRPHL;A;GEO |
Significance: 0.000102845 |
Significance: 0.00020014 |
| ES;J1;A;CL |
Significance: 0.00501471 |
Significance: 0.0341425 |
| ES;E14;A;GEO |
Significance: 0.00113418 |
Significance: 0.00330873 |
| ES;F1216;A;GEO |
Significance: 0.00599407 |
Significance: 0.027423700000000002 |
| ES;v6.5;A;GEO |
Significance: 0.00123011 |
Significance: 0.05662999999999999 |
| Mammary_GLND;A;GEO |
Significance: 0.0430116 |
Significance: 0.050696899999999996 |
| RTN;A;GEO |
Significance: 0.000442432 |
Significance: 0.0144425 |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 5.11526e-05 |
Significance: 0.0169805 |
| ILC2+IL33+IL25;A;GEO |
Significance: 0.00200175 |
Significance: 0.0015831 |
| ILC1;A;GEO |
Significance: 0.00427185 |
Significance: 0.000216464 |
| ILC2;A;GEO |
Significance: 0.0014943 |
Significance: 0.000348713 |
| ILC3;A;GEO |
Significance: 0.00228728 |
Significance: 0.00196687 |
| ILC1_AbxGRMFree;A;GEO |
Significance: 0.000878792 |
Significance: 8.9534e-05 |
| ILC2_AbxGRMFree;A;GEO |
Significance: 0.00358667 |
Significance: 0.00132738 |
| ILC3_AbxGRMFree;A;GEO |
Significance: 0.00262211 |
Significance: 0.000238395 |
| naiveT;CD8+;A;GEO |
Significance: 0.0013469 |
Significance: 0.000538408 |
| T_LCMV;CD8+;D8;A;GEO |
Significance: 0.00227644 |
Significance: 0.0012068 |
| T_LCMV;CD8+;D33;A;GEO |
Significance: 0.00187789 |
Significance: 0.00048733 |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.00265057 |
Significance: 0.0009019739999999999 |
| TEff_SPLND;CD8+;A;GEO |
Significance: 0.0013952 |
Significance: 0.00097905 |
| TExh_SPLND;CD8+;A;GEO |
Significance: 0.00183707 |
Significance: 0.00034176300000000003 |
| TSLEC_SPLND;CD8+A;GEO |
Significance: 0.0062345 |
Significance: 0.000891412 |
| naiveT;trnsgncTCR_SV40-I_CD8;A;GEO |
Significance: 0.00263347 |
Significance: 0.00216345 |
| Teff;TCR_SV40-I_CD8+;D5;A;GEO |
Significance: 0.015572899999999999 |
Significance: 0.00281602 |
| Teff;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.00266147 |
Significance: 0.000942092 |
| cntrl_memT;TCR_SV40-I_CD8+;A;GEO |
Significance: 0.0044502 |
Significance: 0.00214255 |
| T_MLD;TCR_SV40-I_CD8+;D5;A;GEO |
Significance: 0.00202592 |
Significance: 0.000525506 |
| T_MLD;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.000980122 |
Significance: 0.00066919 |
| T_MLD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.00554515 |
Significance: 0.000821355 |
| T_MLD;TCR_SV40-I_CD8+;D21;A;GEO |
Significance: 0.00210209 |
Significance: 0.0096593 |
| T_MLD;TCR_SV40-I_CD8+;D28;A;GEO |
Significance: 0.00502178 |
Significance: 0.000168868 |
| T_MLD;TCR_SV40-I_CD8+;D35;A;GEO |
Significance: 0.00229569 |
Significance: 0.00869989 |
| T_MLD;TCR_SV40-I_CD8+;D60;A;GEO |
Significance: 0.00322948 |
Significance: 0.00160074 |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.00331157 |
Significance: 1.4270799999999999e-05 |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.00201324 |
Significance: 0.000811467 |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D35;A;GEO |
Significance: 0.00553318 |
Significance: 0.0102451 |
| memT_tmrD;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.00252505 |
Significance: 0.000385706 |
| memT_tmrD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.014252 |
Significance: 0.00034853699999999995 |
| iNKT;A;GEO |
Significance: 0.00543676 |
Significance: 0.00469507 |
| aB;A;GEO |
Significance: 0.00156076 |
Significance: 0.000760728 |
| prePLSM_B;A;GEO |
Significance: 0.000511026 |
Significance: 0.00082498 |
| B_SPLND;A;GEO |
Significance: 0.00025179099999999996 |
Significance: 0.00114294 |
| preB_BMD;A;GEO |
Significance: 0.0487823 |
Significance: 0.00231054 |
| B;B220_CD43;A;GEO |
Significance: 0.00129385 |
Significance: 0.00197936 |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.015721799999999998 |
Significance: 0.00121536 |
| MCRPHG_BMD.LPD_Stmltd;A;GEO |
Significance: 0.00023904900000000002 |
Significance: 0.0188292 |
| Erythroid;Ter119+;A;GEO |
Significance: 8.513e-05 |
Significance: 0.00267705 |
| AML;AE9a;A;GEO |
Significance: 0.000507188 |
Significance: 0.00014575799999999998 |
| DC_LPS;6h;A;GEO |
Significance: 0.000309421 |
Significance: 0.0577356 |
| DC_LPS;2h;A;GEO |
Significance: 0.00106587 |
Significance: 0.0414084 |
| DC_pBMD;A;GEO |
Significance: 0.0468533 |
Significance: 0.029622100000000002 |
| ENDRM_ESD;A;GEO |
Significance: 0.000320271 |
Significance: 0.0107265 |
| ROD_PHT_RCPTR;A;GEO |
Significance: 0.00047102 |
Significance: 0.00829214 |
| ROD_PHT_RCPTR_RTN;A;GEO |
Significance: 0.000437928 |
Significance: 0.0181322 |
| GCN_PHT_RCPTR;A;GEO |
Significance: 0.000519547 |
Significance: 0.013614 |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.00039528400000000003 |
Significance: 0.00611929 |
| CN_PHT_RCPTR;A;GEO |
Significance: 0.000628733 |
Significance: 0.00105064 |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.00100138 |
Significance: 0.026909199999999998 |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0006208990000000001 |
Significance: 0.011155 |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.000182888 |
Significance: 0.0100299 |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.000642868 |
Significance: 0.0114517 |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.00340767 |
Significance: 0.00623223 |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00356066 |
Significance: 0.00695104 |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.000847843 |
Significance: 0.010873200000000001 |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.0011484000000000002 |
Significance: 0.00643292 |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.00453189 |
Significance: 0.0270205 |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.000908627 |
Significance: 0.00929963 |
| SKN_EPTHL;A;GEO |
Significance: 0.00440783 |
Significance: 0.00690314 |
| BCK_SKN;A;GEO |
Significance: 0.00560094 |
Significance: 0.0571744 |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 0.00347016 |
Significance: 0.057080700000000005 |
| NPC;G4;A;GEO |
Significance: 0.00131549 |
Significance: 0.012649500000000001 |
| NPC;Ch13;A;GEO |
Significance: 0.000813378 |
Significance: 0.0234349 |
| ePyramidal_NRN;A;GEO |
Significance: 0.00412801 |
Significance: 0.024600099999999996 |
| eCortical_NRN;A;GEO |
Significance: 9.14243e-05 |
Significance: 0.054636699999999996 |
| G_NRN_P;A;GEO |
Significance: 0.00122766 |
Significance: 0.016632 |
| wES;GRM;E16.5;A;GEO |
Significance: 0.000555453 |
Significance: 0.00767232 |
| ThMS_MEC;Aire++;A;GEO |
Significance: 3.56068e-05 |
Significance: 0.0166875 |
| Corticotrope;A;GEO |
Significance: 0.00529571 |
Significance: 0.016291299999999998 |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 0.000322302 |
Significance: 0.0564226 |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.00187067 |
Significance: 0.00289276 |
| pMeg;A;En |
Significance: 8.04643e-05 |
Significance: 0.0157285 |
| EPP;A;En |
Significance: 0.00036994199999999995 |
Significance: 0.00949542 |
| MegEP;A;CL |
Significance: 2.5770999999999998e-05 |
Significance: 0.000891115 |
| pErythroblast;A;En |
Significance: 0.000817659 |
Significance: 0.00535751 |
| MegP;A;En |
Significance: 0.042968099999999995 |
Significance: 0.0248174 |
| pCMP;A;CL |
Significance: 0.0136084 |
Significance: 0.00693804 |
| DC_LPS;4h;A;En |
Significance: 0.000141364 |
Significance: 0.0425214 |
| pGMP;A;En |
Significance: 0.00017962099999999998 |
Significance: 0.020494099999999998 |
| DC;A;En |
Significance: 8.32725e-05 |
Significance: 0.0174599 |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.000138059 |
Significance: 0.019549599999999997 |
| LVR;8wks;D;GEO |
Significance: 0.0007006769999999999 |
Significance: 0.0174231 |
| LNG;8wks;D;GEO |
Significance: 0.00580644 |
Significance: 9.21158e-05 |
| 3134_mammaryADNCRCNM;D;GE |
Significance: 0.00175699 |
Significance: 0.028748900000000004 |
| T_SPLND_blstCLTRD;CD4+;D;GEO |
Significance: 0.00212821 |
Significance: 0.00203285 |
| naiveT_SPLND;CD4+;D;GEO |
Significance: 0.00141516 |
Significance: 0.00073374 |
| Treg;CD4+_CD25+;D;GEO |
Significance: 0.0103833 |
Significance: 0.00018366099999999998 |
| apCD4Eff;D;GEO |
Significance: 0.0020678000000000003 |
Significance: 0.000225761 |
| apTreg;D;GEO |
Significance: 0.00355199 |
Significance: 0.000517533 |
| immatureB_SPLND;CD43-_CD11b-;D;GEO |
Significance: 0.0241391 |
Significance: 6.58593e-05 |
| 416B_MP;CD34+;D;GEO |
Significance: 0.000730399 |
Significance: 7.17529e-06 |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.00482896 |
Significance: 0.0129267 |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 0.03353219999999999 |
Significance: 0.030895 |
| ES;C57BL_6J;R1;D;En |
Significance: 0.00439958 |
Significance: 0.051369399999999996 |
| NIH3T3_FBROBLST;D;En |
Significance: 0.000157166 |
Significance: 0.021137799999999998 |
| 3T3-L1;FBRBLST;D;En |
Significance: 0.000662888 |
Significance: 0.0257621 |
| KDNY;P0;D;En |
Significance: 0.000943725 |
Significance: 0.018475799999999997 |
| KDNY;8wks;D;En |
Significance: 0.0005827240000000001 |
Significance: 6.40655e-05 |
| LVR;8wks;D;En |
Significance: 0.00298281 |
Significance: 0.0109565 |
| wES;LVR;E11.5;D.;En |
Significance: 0.00166359 |
Significance: 0.00776743 |
| LVR;P0;D;En |
Significance: 0.000244482 |
Significance: 0.019737900000000003 |
| LNG;8wks;D;En |
Significance: 0.0009688260000000001 |
Significance: 0.00010384200000000001 |
| LNG;P0;D;En |
Significance: 0.00165679 |
Significance: 1.5475399999999998e-06 |
| wES;LNG;E14.5;D;En |
Significance: 0.000714875 |
Significance: 0.00240292 |
| Heart;8wks;D;En |
Significance: 0.000456839 |
Significance: 2.2126999999999997e-05 |
| Heart;P0;D;En |
Significance: 0.000426404 |
Significance: 0.00046269199999999995 |
| Adipose;D;En |
Significance: 0.00564369 |
Significance: 0.0109682 |
| RTN;D1;D;En |
Significance: 0.00474486 |
Significance: 0.0150183 |
| wES;RTN;E11.5;D;En |
Significance: 0.00391807 |
Significance: 0.000482658 |
| CRBLM;D;En |
Significance: 0.000157388 |
Significance: 0.00939816 |
| apThEff;CD4+;8wks;D;En. |
Significance: 0.00232394 |
Significance: 0.000429456 |
| abTreg;CD4+_CD25+;8wks;D;En |
Significance: 0.00490117 |
Significance: 0.000544317 |
| apTreg;8wk;D;En |
Significance: 0.00652665 |
Significance: 0.00020406900000000003 |
| EP;c-kit+CD71+Ter119+;D;En |
Significance: 0.00030661400000000003 |
Significance: 0.00786953 |
| 416B_MP;CD34+;D;En |
Significance: 0.0013545999999999999 |
Significance: 0.00017041 |
| Fr_BRN;P0;D;En |
Significance: 0.00530872 |
Significance: 0.0251672 |
| Hnd_BRN;P0;D;En |
Significance: 0.00150235 |
Significance: 0.057226400000000004 |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.00810559 |
Significance: 0.0112384 |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.00134518 |
Significance: 0.023575299999999997 |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.00244214 |
Significance: 0.022649400000000004 |
| Md_BRN;D;En |
Significance: 0.00128388 |
Significance: 0.024003599999999996 |
| WHL_BRN;8wks;D;En |
Significance: 4.06759e-05 |
Significance: 0.0192147 |
| wES;Telenchephalon;D;En |
Significance: 0.00105485 |
Significance: 0.021111599999999998 |
| wES;limb;E11.5;D;En |
Significance: 0.000389021 |
Significance: 0.00195927 |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.00111406 |
Significance: 0.0371234 |
| wES;FaceP;E14.5;D;En |
Significance: 1.25629e-05 |
Significance: 0.0231723 |
| LRG_INTSTN;8wks;D;En |
Significance: 0.00547929 |
Significance: 0.00423472 |
| STMCH;P0;D;En |
Significance: 0.00034766800000000003 |
Significance: 0.00283451 |
| SKLTL_Muscle;8wks;D;En |
Significance: 2.82545e-05 |
Significance: 0.000992944 |
| Spleen;8wks;D;En |
Significance: 0.00267978 |
Significance: 7.570359999999999e-05 |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.000770112 |
Significance: 0.00289281 |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.000410828 |
Significance: 8.233959999999999e-05 |
| A20;B-lymphoma;D;En |
Significance: 0.00136125 |
Significance: 0.00119128 |
| AML;D;En |
Significance: 0.054608300000000005 |
Significance: 0.0317825 |
| CH12.LX;D;En |
Significance: 0.047510500000000004 |
Significance: 0.00022665 |
| MEF.GATA-1-ER;D;En |
Significance: 0.0004911059999999999 |
Significance: 0.0503235 |
| NIH3T3;D;En |
Significance: 0.000251445 |
Significance: 0.00260182 |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.000818696 |
Significance: 0.00932866 |
| MN1;D;En |
Significance: 0.00151501 |
Significance: 0.00569704 |
| 3T3-L1;D;En |
Significance: 0.000725442 |
Significance: 0.015312899999999999 |
Unique Cell types only TF-MoDISco finds Elk4: #31
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En |
Significance: 2.1360500000000003e-05 | Absent |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.00051312 | Absent |
| wES;FaceP;E11.5;D;En |
Significance: 0.000600249 | Absent |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.000399893 | Absent |
| MEL;D;En |
Significance: 0.000194952 | Absent |
| MEL_Znc_di-cl;24h;D;En |
Significance: 0.054394000000000005 | Absent |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.000807103 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.000520242 | Absent |
| NPC;F129;A;GEO |
Significance: 0.000855563 | Absent |
| NPC;mE6;A;GEO |
Significance: 0.000952295 | Absent |
| iPV_NRN;A;GEO |
Significance: 0.00494736 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.00655819 | Absent |
| ES;E14;A;CL |
Significance: 0.000610425 | Absent |
| Melanotrope;A;GEO |
Significance: 0.00241891 | Absent |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.00313728 | Absent |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.00357853 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.00415213 | Absent |
| Mammary_BptfKO;A;GEO |
Significance: 0.00329885 | Absent |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.00403346 | Absent |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.00491139 | Absent |
| ES;D0;D;GEO |
Significance: 0.0035544000000000005 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.0009569139999999999 | Absent |
| ES;E14;D;En |
Significance: 0.00598095 | Absent |
| FBRBLST;8wks;D;En |
Significance: 0.0009181889999999999 | Absent |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.00383896 | Absent |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 0.00142003 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 0.00044207599999999996 | Absent |
| Muller;P12;D;En |
Significance: 0.000170656 | Absent |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.00017277 | Absent |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.00583387 | Absent |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.0009225310000000001 | Absent |
Unique Cell types only Homer finds Elk4: #2
| Cell type | Modisco | Homer |
|---|---|---|
| immatureB_SPLND;CD43-_CD11b-;D;En | Absent |
Significance: 0.00019988900000000002 |
| proB;A;CL | Absent |
Significance: 0.000277306 |