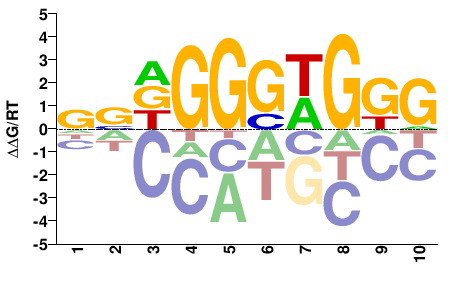
# Parameters
data_name = "Sall4"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:33:25,783 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Sall4: #27
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E11.5;D;En |
Significance: 0.0539216 |
Significance: 0.011448 |
| wES;FaceP;E11.5;D;En |
Significance: 0.042638800000000004 |
Significance: 0.0169216 |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.031052499999999997 |
Significance: 0.021325099999999996 |
| plasmaB;A;CL |
Significance: 0.034224199999999996 |
Significance: 0.0117914 |
| aB_LSP+a_d_dextran;A;CL |
Significance: 0.0587281 |
Significance: 0.0391416 |
| Mast;A;CL |
Significance: 0.051020300000000005 |
Significance: 0.0191998 |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.033016699999999996 |
Significance: 0.00204017 |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.0191459 |
Significance: 0.039908099999999995 |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0293733 |
Significance: 0.0523976 |
| CD4+CD8-TCR+CD69+CD24+CD25-;B2defM;A;CL |
Significance: 0.0342025 |
Significance: 0.0481712 |
| CD4+CD8-TCR+CD69-CD24-CD25-;B2defM;A;CL |
Significance: 0.0523688 |
Significance: 0.0399691 |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.0397779 |
Significance: 0.0144284 |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.019745500000000003 |
Significance: 0.0146886 |
| DP;A;CL |
Significance: 0.048693400000000005 |
Significance: 0.051097199999999995 |
| iVIP_NRN;A;GEO |
Significance: 0.049015300000000005 |
Significance: 0.0166756 |
| BRN;FRNTL_CRTX;A;CL |
Significance: 0.0236669 |
Significance: 0.05536669999999999 |
| BRN;CRBLM;A;CL |
Significance: 0.0592933 |
Significance: 0.0463485 |
| LNG;8wks;D;GEO |
Significance: 0.0409189 |
Significance: 0.0237889 |
| naiveT;trnsgncTCR_SV40-I_CD8;A;GEO |
Significance: 0.031027 |
Significance: 0.00678374 |
| KDNY;P0;D;En |
Significance: 0.027595099999999997 |
Significance: 0.029698400000000003 |
| LNG;P0;D;En |
Significance: 0.00401458 |
Significance: 0.014107499999999999 |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.0480327 |
Significance: 0.00321338 |
| Heart;P0;D;En |
Significance: 0.018453 |
Significance: 0.0182362 |
| RTN;A;GEO |
Significance: 0.052200800000000006 |
Significance: 0.0203412 |
| RTN;D1;D;En |
Significance: 0.0474592 |
Significance: 0.0165382 |
| wES;MSDRM;E11.5;D;En |
Significance: 0.0252928 |
Significance: 0.00884917 |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.03652709999999999 |
Significance: 0.0595308 |
Unique Cell types only TF-MoDISco finds Sall4: #119
| Cell type | Modisco | Homer |
|---|---|---|
| memT;CD4+;A;CL |
Significance: 0.0242813 | Absent |
| NK_SPLND;A;CL |
Significance: 0.00768415 | Absent |
| preB;A;CL |
Significance: 0.05821799999999999 | Absent |
| immatureB;A;CL |
Significance: 0.026057099999999996 | Absent |
| matureB;A;CL |
Significance: 0.04999 | Absent |
| B1_B;A;CL |
Significance: 0.059148 | Absent |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 0.0501776 | Absent |
| ESNPHL_SPLND_BlbCA;CL |
Significance: 0.0420953 | Absent |
| Tfh_LCMV;A;CL |
Significance: 0.022333000000000002 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.044839699999999996 | Absent |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.014161400000000001 | Absent |
| naiveT_mut;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.0284075 | Absent |
| largeDPprecrsr;CD4+CD8+CD69-;A;CL |
Significance: 0.0409485 | Absent |
| CD4+CD8intTCR+CD69+CD24+;MHC2defM;A;CL |
Significance: 0.052488 | Absent |
| CD4+CD8+TCR+CD69+CD24+;MHC2defM;A;C |
Significance: 0.05813430000000001 | Absent |
| CD4+CD8+TCR+CD69-CD24-;MHC2defM;A;CL |
Significance: 0.00207168 | Absent |
| CD4+CD8-TCR+CD25+Foxp3-;A;CL |
Significance: 0.0243273 | Absent |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL |
Significance: 0.0371866 | Absent |
| Erythroblast;A;CL |
Significance: 0.04176130000000001 | Absent |
| ISP;A;CL |
Significance: 0.0455498 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.024400400000000003 | Absent |
| Heart;D;En |
Significance: 0.0522419 | Absent |
| Adipose;A;CL |
Significance: 0.05615359999999999 | Absent |
| Mammary;A;CL |
Significance: 0.0425613 | Absent |
| placenta;A;CL |
Significance: 0.03995580000000001 | Absent |
| STMCH;A;CL |
Significance: 0.030782599999999997 | Absent |
| PNCRS;A;CL |
Significance: 0.032515800000000004 | Absent |
| eye-NRL_RTN;A;CL |
Significance: 0.0194633 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.030100799999999997 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.05344550000000001 | Absent |
| BRN;MDL_OBLNGT;A;CL |
Significance: 0.0407647 | Absent |
| restingB;A;CL |
Significance: 0.018993299999999998 | Absent |
| aB;CD180;A;CL |
Significance: 0.0487091 | Absent |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.05434119999999999 | Absent |
| ILC2precrsr;A;CL |
Significance: 0.028307 | Absent |
| NCR+ILC3;A;CL |
Significance: 0.0393698 | Absent |
| ILC2_Nippo;A;CL |
Significance: 0.035087400000000005 | Absent |
| ILC2;A;CL |
Significance: 0.045546300000000005 | Absent |
| wCH12;A;CL |
Significance: 0.041140300000000005 | Absent |
| CH12;med14;A;CL |
Significance: 0.0560816 | Absent |
| wCH12_drug;A;CL |
Significance: 0.052342999999999994 | Absent |
| NK_LVRD;CD19-CD3e-NKp46+DX+Trail-;A;GEO |
Significance: 0.040519 | Absent |
| ES;J1;A;CL |
Significance: 0.0146666 | Absent |
| ES;E14;A;GEO |
Significance: 0.0360416 | Absent |
| ES;F1216;A;GEO |
Significance: 0.030896499999999997 | Absent |
| ES;v6.5;A;GEO |
Significance: 0.0404508 | Absent |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.042243 | Absent |
| T_LCMV;CD8+;D8;A;GEO |
Significance: 0.0501778 | Absent |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.0362851 | Absent |
| TSLEC_SPLND;CD8+A;GEO |
Significance: 0.0187221 | Absent |
| cntrl_memT;TCR_SV40-I_CD8+;A;GEO |
Significance: 0.0529547 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D5;A;GEO |
Significance: 0.00283663 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.0200699 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D21;A;GEO |
Significance: 0.014206899999999998 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D35;A;GEO |
Significance: 0.0458579 | Absent |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.0328776 | Absent |
| preB_BMD;A;GEO |
Significance: 0.026958600000000003 | Absent |
| Erythroid;Ter119+;A;GEO |
Significance: 0.044337699999999994 | Absent |
| AML;AE9a;A;GEO |
Significance: 0.029004200000000004 | Absent |
| ENDRM_ESD;A;GEO |
Significance: 0.0537149 | Absent |
| ROD_PHT_RCPTR;A;GEO |
Significance: 0.0310373 | Absent |
| GCN_PHT_RCPTR;A;GEO |
Significance: 0.0261038 | Absent |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.0591423 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.0260192 | Absent |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0530413 | Absent |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0501871 | Absent |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.0250166 | Absent |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.045787699999999994 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.048026 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.0298449 | Absent |
| SKN_EPTHL;A;GEO |
Significance: 0.000941148 | Absent |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 0.0507014 | Absent |
| NPC;G4;A;GEO |
Significance: 0.0593288 | Absent |
| NPC;Ch13;A;GEO |
Significance: 0.056262900000000005 | Absent |
| NPC;mE6;A;GEO |
Significance: 0.048688 | Absent |
| iPV_NRN;A;GEO |
Significance: 0.0301019 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.0573289 | Absent |
| wES;GRM;E16.5;A;GEO |
Significance: 0.045513599999999994 | Absent |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.00698779 | Absent |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.00460025 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.00313551 | Absent |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 0.05494299999999999 | Absent |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.00391712 | Absent |
| Mammary_BptfKO;A;GEO |
Significance: 0.038860900000000004 | Absent |
| EPP;A;En |
Significance: 0.0239784 | Absent |
| MegEP;A;CL |
Significance: 0.00728376 | Absent |
| pErythroblast;A;En |
Significance: 0.0381141 | Absent |
| DC_LPS;4h;A;En |
Significance: 0.044996499999999995 | Absent |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.00464249 | Absent |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.013112 | Absent |
| ES;D0;D;GEO |
Significance: 0.0182037 | Absent |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.0449566 | Absent |
| apCD4Eff;D;GEO |
Significance: 0.0561155 | Absent |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.0334659 | Absent |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 0.0231263 | Absent |
| ES;E14;D;En |
Significance: 0.0216807 | Absent |
| ES;C57BL_6J;R1;D;En |
Significance: 0.00183521 | Absent |
| 3T3-L1;FBRBLST;D;En |
Significance: 0.0281101 | Absent |
| wES;LVR;E11.5;D.;En |
Significance: 0.047122500000000005 | Absent |
| wES;RTN;E11.5;D;En |
Significance: 0.0232159 | Absent |
| Muller;P12;D;En |
Significance: 0.0511003 | Absent |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.021902200000000004 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.0383452 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.016668099999999998 | Absent |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.0457448 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.037967900000000006 | Absent |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.0265856 | Absent |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.0209622 | Absent |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.0356272 | Absent |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.025400799999999998 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.00586878 | Absent |
| LRG_INTSTN;8wks;D;En |
Significance: 0.0319474 | Absent |
| STMCH;P0;D;En |
Significance: 0.0354161 | Absent |
| SKLTL_Muscle;8wks;D;En |
Significance: 0.014351599999999999 | Absent |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.044360500000000004 | Absent |
| CH12.LX;D;En |
Significance: 0.057367499999999995 | Absent |
| NIH3T3;D;En |
Significance: 0.038844699999999996 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.0130631 | Absent |
| MN1;D;En |
Significance: 0.043125 | Absent |
Unique Cell types only Homer finds Sall4: #24
| Cell type | Modisco | Homer |
|---|---|---|
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D35;A;GEO | Absent |
Significance: 0.0325527 |
| 416B;HMTP_SPNSN;CD34+;D;En | Absent |
Significance: 0.0196984 |
| aB_LPS;A;CL | Absent |
Significance: 0.0296094 |
| aB_LPS+IL4;A;CL | Absent |
Significance: 0.0565388 |
| MCRPHG_BMD.LPD_Stmltd;A;GEO | Absent |
Significance: 0.0237435 |
| B;B220_CD43;A;GEO | Absent |
Significance: 0.0312223 |
| Tfh_mut;A;CL | Absent |
Significance: 0.0203388 |
| Tfr;CD4+CD44+CXCR5+PD1+CD25+;A;CL | Absent |
Significance: 0.0329846 |
| naiveT;CD4+_CD44-_CD62L-_CD25-;A;CL | Absent |
Significance: 0.0476142 |
| ePyramidal_NRN;A;GEO | Absent |
Significance: 0.043745400000000004 |
| Adrenal;A;CL | Absent |
Significance: 0.011158 |
| pMeg;A;En | Absent |
Significance: 0.040405800000000006 |
| MegP;A;En | Absent |
Significance: 0.0174955 |
| pCMP;A;CL | Absent |
Significance: 0.00925723 |
| pGMP;A;En | Absent |
Significance: 0.046213800000000006 |
| BRN;PTMN;A;CL | Absent |
Significance: 0.00429661 |
| BRN;SNSR_MTR_CRTX;A;CL | Absent |
Significance: 0.024488 |
| LVR;8wks;D;GEO | Absent |
Significance: 0.021620300000000002 |
| LNG_TRM_HSC;A;CL | Absent |
Significance: 0.0179385 |
| KDNY;8wks;D;En | Absent |
Significance: 0.0106806 |
| LNG;8wks;D;En | Absent |
Significance: 0.026038299999999997 |
| ILC3;A;GEO | Absent |
Significance: 0.050218 |
| T_MLD;TCR_SV40-I_CD8+;D14;A;GEO | Absent |
Significance: 0.00523222 |
| WHL_BRN;8wks;D;En | Absent |
Significance: 0.0369105 |