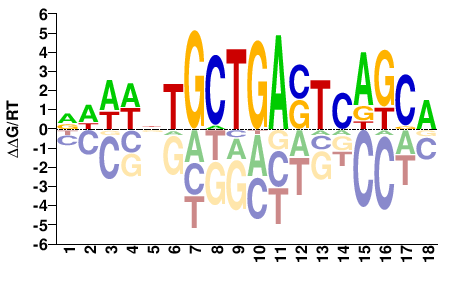
# Parameters
data_name = "Mafg"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:18:43,111 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Mafg: #31
| Cell type | Modisco | Homer |
|---|---|---|
| memT;CD4+;A;CL |
Significance: 0.05930509999999999 |
Significance: 0.04918959999999999 |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.0451033 |
Significance: 0.020680599999999997 |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.0231412 |
Significance: 0.000552581 |
| AML;D;En |
Significance: 0.00246633 |
Significance: 0.0337704 |
| Erythroid;Ter119+;A;GEO |
Significance: 0.00238802 |
Significance: 2.90343e-06 |
| AML;AE9a;A;GEO |
Significance: 0.00325726 |
Significance: 0.000163762 |
| MEF.GATA-1-ER;D;En |
Significance: 0.00452942 |
Significance: 0.00028213 |
| NIH3T3;D;En |
Significance: 0.0319087 |
Significance: 0.0139553 |
| MN1;D;En |
Significance: 0.0571012 |
Significance: 0.026365800000000002 |
| 3T3-L1;D;En |
Significance: 0.0356457 |
Significance: 0.0081779 |
| MCRPHG;A;CL |
Significance: 0.021782 |
Significance: 0.00213735 |
| Erythroblast;A;CL |
Significance: 0.00231423 |
Significance: 0.00881238 |
| iPV_NRN;A;GEO |
Significance: 0.0207867 |
Significance: 0.0199348 |
| EPP;A;En |
Significance: 0.00174608 |
Significance: 0.00150825 |
| pErythroblast;A;En |
Significance: 0.0039585 |
Significance: 0.024134700000000002 |
| MegP;A;En |
Significance: 0.00317644 |
Significance: 0.00710012 |
| pCMP;A;CL |
Significance: 0.00298493 |
Significance: 0.0130762 |
| DC;A;En |
Significance: 0.0473136 |
Significance: 0.00384867 |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.00162529 |
Significance: 9.072649999999999e-05 |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.00605384 |
Significance: 0.0110146 |
| 3134_mammaryADNCRCNM;D;GE |
Significance: 0.04530580000000001 |
Significance: 0.00955507 |
| gamma_delta_TCR;A;CL |
Significance: 0.0475812 |
Significance: 0.0201894 |
| NIH3T3_FBROBLST;D;En |
Significance: 0.0522259 |
Significance: 0.0239524 |
| wES;LVR;E11.5;D.;En |
Significance: 0.00295686 |
Significance: 0.00366605 |
| ES;v6.5;A;GEO |
Significance: 0.00613149 |
Significance: 0.0198133 |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 0.0031854 |
Significance: 0.0347902 |
| Adipose;D;En |
Significance: 0.0380615 |
Significance: 0.027754900000000002 |
| EP;c-kit+CD71+Ter119+;D;En |
Significance: 0.00334552 |
Significance: 0.00213668 |
| 416B_MP;CD34+;D;En |
Significance: 0.00971224 |
Significance: 0.00141534 |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.0597793 |
Significance: 0.0371071 |
| TSLEC_SPLND;CD8+A;GEO |
Significance: 0.0591026 |
Significance: 0.0465827 |
Unique Cell types only TF-MoDISco finds Mafg: #74
| Cell type | Modisco | Homer |
|---|---|---|
| MPP;A;CL |
Significance: 0.0026438000000000004 | Absent |
| Th17;CD4+;A;CL |
Significance: 0.029992599999999998 | Absent |
| proB;A;CL |
Significance: 0.00492515 | Absent |
| matureB;A;CL |
Significance: 0.0318 | Absent |
| iNKT;A;GEO |
Significance: 0.0243945 | Absent |
| SKLTL_Muscle;8wks;D;En |
Significance: 0.0146459 | Absent |
| plasmaB;A;CL |
Significance: 0.00611205 | Absent |
| aB_LPS;A;CL |
Significance: 0.011947200000000002 | Absent |
| B;B220_CD43;A;GEO |
Significance: 0.00331674 | Absent |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.043711 | Absent |
| aB_LSP+a_d_dextran;A;CL |
Significance: 0.053940300000000004 | Absent |
| CH12.LX;D;En |
Significance: 0.0489441 | Absent |
| Mast;A;CL |
Significance: 0.00531762 | Absent |
| MEL;D;En |
Significance: 0.00248318 | Absent |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 0.00509254 | Absent |
| ESNPHL_SPLND_BlbCA;CL |
Significance: 0.01567 | Absent |
| NTRPHL;A;CL |
Significance: 0.0423286 | Absent |
| Tfh_LCMV;A;CL |
Significance: 0.028355 | Absent |
| Tfh;A;CL |
Significance: 0.027179000000000002 | Absent |
| ENDRM_ESD;A;GEO |
Significance: 0.00333697 | Absent |
| GCN_PHT_RCPTR;A;GEO |
Significance: 0.056432500000000003 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.0352216 | Absent |
| Tfr;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.0583521 | Absent |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.0452933 | Absent |
| smallDPprecrsor;CD4+CD8+CD69-;A;CL |
Significance: 0.00109533 | Absent |
| NPC;F129;A;GEO |
Significance: 0.0578001 | Absent |
| DC_BMD;A;CL |
Significance: 0.015480700000000002 | Absent |
| NPC;G4;A;GEO |
Significance: 0.00510406 | Absent |
| DP3a;A;CL |
Significance: 0.000971199 | Absent |
| ES;E14;A;CL |
Significance: 3.15649e-05 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.0477176 | Absent |
| LNG;A;CL |
Significance: 0.0549974 | Absent |
| Muscle;A;CL |
Significance: 9.211959999999999e-05 | Absent |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.00253978 | Absent |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.00557509 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.00255834 | Absent |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 0.0001356 | Absent |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.000261004 | Absent |
| pMeg;A;En |
Significance: 0.00237216 | Absent |
| MegEP;A;CL |
Significance: 0.00278462 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.00781758 | Absent |
| DC_LPS;4h;A;En |
Significance: 0.026712299999999998 | Absent |
| pGMP;A;En |
Significance: 0.0020012999999999997 | Absent |
| BRN;MDL_OBLNGT;A;CL |
Significance: 0.0118125 | Absent |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.00155395 | Absent |
| BRN;CRBLM;A;CL |
Significance: 0.00174108 | Absent |
| ES;D0;D;GEO |
Significance: 0.000320133 | Absent |
| LVR;8wks;D;GEO |
Significance: 4.48091e-05 | Absent |
| LNG_TRM_HSC;A;CL |
Significance: 0.00245786 | Absent |
| SHRT_TRM_HSC;A;CL |
Significance: 0.00210276 | Absent |
| aB_LPS+IL4_24h;A;CL |
Significance: 0.041411699999999996 | Absent |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 0.00147196 | Absent |
| CH12_QNTPLE;A;CL |
Significance: 0.00168388 | Absent |
| wCH12;A;CL |
Significance: 0.0559303 | Absent |
| CH12;med14;A;CL |
Significance: 0.037025999999999996 | Absent |
| ES;E14;D;En |
Significance: 0.00110671 | Absent |
| CLP;A;CL |
Significance: 0.00576895 | Absent |
| ES;C57BL_6J;R1;D;En |
Significance: 0.00188328 | Absent |
| KDNY;8wks;D;En |
Significance: 7.58322e-06 | Absent |
| NTRPHL;A;GEO |
Significance: 0.00225196 | Absent |
| ES;J1;A;CL |
Significance: 0.00655503 | Absent |
| ES;E14;A;GEO |
Significance: 0.027085700000000004 | Absent |
| ES;F1216;A;GEO |
Significance: 0.014095600000000002 | Absent |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.00321312 | Absent |
| LVR;8wks;D;En |
Significance: 0.00492211 | Absent |
| LVR;P0;D;En |
Significance: 0.0007475110000000001 | Absent |
| wES;LNG;E14.5;D;En |
Significance: 0.010805799999999999 | Absent |
| MEL_Znc_di-cl;24h;D;En |
Significance: 0.00151132 | Absent |
| ILC2;A;GEO |
Significance: 0.05531559999999999 | Absent |
| ILC3;A;GEO |
Significance: 0.0157659 | Absent |
| ILC1_AbxGRMFree;A;GEO |
Significance: 0.0462651 | Absent |
| ILC2_AbxGRMFree;A;GEO |
Significance: 0.0295295 | Absent |
| CRBLM;D;En |
Significance: 0.0211918 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.00599626 | Absent |
Unique Cell types only Homer finds Mafg: #28
| Cell type | Modisco | Homer |
|---|---|---|
| STMCH;P0;D;En | Absent |
Significance: 0.0490592 |
| MCRPHG_BMD.LPD_Stmltd;A;GEO | Absent |
Significance: 0.0217103 |
| BSPHL;A;CL | Absent |
Significance: 0.00735854 |
| DC_LPS;2h;A;GEO | Absent |
Significance: 0.00137854 |
| MTR_NRN_MN1;methyl3;D;En | Absent |
Significance: 0.0282322 |
| BCK_SKN;A;GEO | Absent |
Significance: 0.0559336 |
| eCortical_NRN;A;GEO | Absent |
Significance: 0.0542778 |
| ES;MEF;A;CL | Absent |
Significance: 0.05379 |
| Adrenal;A;CL | Absent |
Significance: 0.0125256 |
| Mammary_BptfKO;A;GEO | Absent |
Significance: 0.027419799999999998 |
| BRN;OLFTRY_BLB;A;CL | Absent |
Significance: 0.013865 |
| BRN;FRNTL_CRTX;A;CL | Absent |
Significance: 0.05672340000000001 |
| BRN;SNSR_MTR_CRTX;A;CL | Absent |
Significance: 0.0439784 |
| T_SPLND_blstCLTRD;CD4+;D;GEO | Absent |
Significance: 0.0411083 |
| gamma_delta_TCR_M-S-;A;CL | Absent |
Significance: 0.0460639 |
| NCR+ILC3;A;CL | Absent |
Significance: 0.0134761 |
| ILC2_Nippo;A;CL | Absent |
Significance: 0.016896 |
| 416B_MP;CD34+;D;GEO | Absent |
Significance: 0.0161707 |
| NCR+ILC3+IL23;A;CL | Absent |
Significance: 0.00951923 |
| WHL_BRN;8wks;D;GEO | Absent |
Significance: 0.0280057 |
| aB_LPS+IL4+Oligomycin_24h;A;CL | Absent |
Significance: 0.0462535 |
| FBRBLST;8wks;D;En | Absent |
Significance: 0.0102264 |
| wCH12_drug;A;CL | Absent |
Significance: 0.0130711 |
| 3T3-L1;FBRBLST;D;En | Absent |
Significance: 0.0175982 |
| Mammary_GLND;A;GEO | Absent |
Significance: 0.00631311 |
| ILC1;A;GEO | Absent |
Significance: 0.0376569 |
| Muller;P12;D;En | Absent |
Significance: 0.01024 |
| WHL_BRN;8wks;D;En | Absent |
Significance: 0.020016 |