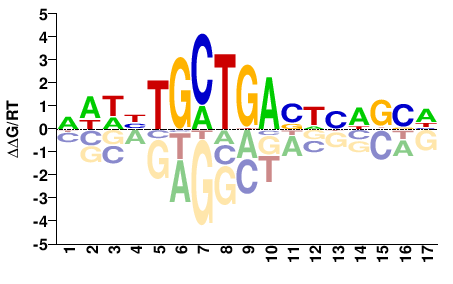
# Parameters
data_name = "Maf"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:18:30,338 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Maf: #27
| Cell type | Modisco | Homer |
|---|---|---|
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.0450045 |
Significance: 0.0223053 |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.0343894 |
Significance: 0.00183497 |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.0477456 |
Significance: 0.0566764 |
| Erythroid;Ter119+;A;GEO |
Significance: 0.00379413 |
Significance: 0.000332155 |
| AML;AE9a;A;GEO |
Significance: 0.00333799 |
Significance: 0.0005185 |
| MEF.GATA-1-ER;D;En |
Significance: 0.00538894 |
Significance: 0.00067673 |
| NIH3T3;D;En |
Significance: 0.0269115 |
Significance: 0.029676400000000002 |
| 3T3-L1;D;En |
Significance: 0.035549800000000006 |
Significance: 0.013293700000000002 |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.031965 |
Significance: 0.054495400000000006 |
| MCRPHG;A;CL |
Significance: 0.0296281 |
Significance: 0.00404352 |
| Erythroblast;A;CL |
Significance: 0.00290288 |
Significance: 0.017098500000000003 |
| iPV_NRN;A;GEO |
Significance: 0.021889 |
Significance: 0.0199348 |
| EPP;A;En |
Significance: 0.00288025 |
Significance: 0.00318701 |
| pErythroblast;A;En |
Significance: 0.0043329 |
Significance: 0.046706300000000006 |
| MegP;A;En |
Significance: 0.00361343 |
Significance: 0.00803526 |
| pCMP;A;CL |
Significance: 0.00374238 |
Significance: 0.01646 |
| DC;A;En |
Significance: 0.0473136 |
Significance: 0.00753353 |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.00270037 |
Significance: 0.000411395 |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.00966191 |
Significance: 0.0240363 |
| 3134_mammaryADNCRCNM;D;GE |
Significance: 0.042944300000000005 |
Significance: 0.019131400000000003 |
| NIH3T3_FBROBLST;D;En |
Significance: 0.0489433 |
Significance: 0.0510328 |
| wES;LVR;E11.5;D.;En |
Significance: 0.00324819 |
Significance: 0.00965684 |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 0.00409651 |
Significance: 0.0466146 |
| Adipose;D;En |
Significance: 0.0380615 |
Significance: 0.039324300000000006 |
| EP;c-kit+CD71+Ter119+;D;En |
Significance: 0.00395443 |
Significance: 0.00497193 |
| 416B_MP;CD34+;D;En |
Significance: 0.012262799999999999 |
Significance: 0.00302298 |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.0592947 |
Significance: 0.058282900000000006 |
Unique Cell types only TF-MoDISco finds Maf: #69
| Cell type | Modisco | Homer |
|---|---|---|
| MPP;A;CL |
Significance: 0.00348686 | Absent |
| Th17;CD4+;A;CL |
Significance: 0.049303 | Absent |
| proB;A;CL |
Significance: 0.00674799 | Absent |
| iNKT;A;GEO |
Significance: 0.037313 | Absent |
| SKLTL_Muscle;8wks;D;En |
Significance: 0.0176942 | Absent |
| plasmaB;A;CL |
Significance: 0.00810399 | Absent |
| aB_LPS;A;CL |
Significance: 0.018482 | Absent |
| B;B220_CD43;A;GEO |
Significance: 0.00838407 | Absent |
| AML;D;En |
Significance: 0.00260292 | Absent |
| CH12.LX;D;En |
Significance: 0.046854 | Absent |
| MEL;D;En |
Significance: 0.0027603000000000003 | Absent |
| Mast;A;CL |
Significance: 0.0059895 | Absent |
| MEL_Znc_di-cl;24h;D;En |
Significance: 0.00285953 | Absent |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 0.00598836 | Absent |
| ESNPHL_SPLND_BlbCA;CL |
Significance: 0.0219362 | Absent |
| NTRPHL;A;CL |
Significance: 0.044366300000000004 | Absent |
| Tfh_LCMV;A;CL |
Significance: 0.050977800000000004 | Absent |
| Tfh;A;CL |
Significance: 0.030817700000000003 | Absent |
| ENDRM_ESD;A;GEO |
Significance: 0.011865700000000002 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.052266999999999994 | Absent |
| Tfr;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.057941099999999995 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.052194000000000004 | Absent |
| smallDPprecrsor;CD4+CD8+CD69-;A;CL |
Significance: 0.00155841 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.0432151 | Absent |
| DC_BMD;A;CL |
Significance: 0.0310365 | Absent |
| NPC;G4;A;GEO |
Significance: 0.014966 | Absent |
| DP3a;A;CL |
Significance: 0.00128097 | Absent |
| ES;E14;A;CL |
Significance: 8.365e-05 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.00808564 | Absent |
| Muscle;A;CL |
Significance: 0.000122771 | Absent |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.00371195 | Absent |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.00848008 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.0035586 | Absent |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 0.000515751 | Absent |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.00046908199999999995 | Absent |
| pMeg;A;En |
Significance: 0.00323815 | Absent |
| MegEP;A;CL |
Significance: 0.00386084 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.010324 | Absent |
| DC_LPS;4h;A;En |
Significance: 0.0341863 | Absent |
| pGMP;A;En |
Significance: 0.00320859 | Absent |
| BRN;MDL_OBLNGT;A;CL |
Significance: 0.013622899999999999 | Absent |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.00186305 | Absent |
| BRN;CRBLM;A;CL |
Significance: 0.00276148 | Absent |
| ES;D0;D;GEO |
Significance: 0.000851938 | Absent |
| LVR;8wks;D;GEO |
Significance: 0.00040637 | Absent |
| LNG_TRM_HSC;A;CL |
Significance: 0.00358278 | Absent |
| SHRT_TRM_HSC;A;CL |
Significance: 0.0029006 | Absent |
| aB_LPS+IL4_24h;A;CL |
Significance: 0.0432555 | Absent |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 0.00227028 | Absent |
| CH12_QNTPLE;A;CL |
Significance: 0.00253157 | Absent |
| wCH12;A;CL |
Significance: 0.0518757 | Absent |
| CH12;med14;A;CL |
Significance: 0.0575552 | Absent |
| ES;E14;D;En |
Significance: 0.00170742 | Absent |
| CLP;A;CL |
Significance: 0.00693319 | Absent |
| ES;C57BL_6J;R1;D;En |
Significance: 0.00296721 | Absent |
| KDNY;8wks;D;En |
Significance: 6.91433e-05 | Absent |
| NTRPHL;A;GEO |
Significance: 0.00257975 | Absent |
| ES;J1;A;CL |
Significance: 0.00792772 | Absent |
| ES;E14;A;GEO |
Significance: 0.0298282 | Absent |
| ES;F1216;A;GEO |
Significance: 0.0379755 | Absent |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.00776373 | Absent |
| ES;v6.5;A;GEO |
Significance: 0.016083 | Absent |
| LVR;8wks;D;En |
Significance: 0.00713199 | Absent |
| LVR;P0;D;En |
Significance: 0.0010488 | Absent |
| wES;LNG;E14.5;D;En |
Significance: 0.0148079 | Absent |
| CRBLM;D;En |
Significance: 0.025704200000000003 | Absent |
| ILC3;A;GEO |
Significance: 0.0237488 | Absent |
| ILC2_AbxGRMFree;A;GEO |
Significance: 0.0319477 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.00732259 | Absent |
Unique Cell types only Homer finds Maf: #24
| Cell type | Modisco | Homer |
|---|---|---|
| MCRPHG_BMD.LPD_Stmltd;A;GEO | Absent |
Significance: 0.05152 |
| BSPHL;A;CL | Absent |
Significance: 0.00735854 |
| DC_LPS;2h;A;GEO | Absent |
Significance: 0.00235876 |
| MN1;D;En | Absent |
Significance: 0.0492029 |
| MTR_NRN_MN1;methyl3;D;En | Absent |
Significance: 0.057108400000000004 |
| Adrenal;A;CL | Absent |
Significance: 0.015582599999999999 |
| Mammary_BptfKO;A;GEO | Absent |
Significance: 0.037096 |
| BRN;OLFTRY_BLB;A;CL | Absent |
Significance: 0.036396 |
| BRN;FRNTL_CRTX;A;CL | Absent |
Significance: 0.057255099999999996 |
| BRN;SNSR_MTR_CRTX;A;CL | Absent |
Significance: 0.0505743 |
| gamma_delta_TCR;A;CL | Absent |
Significance: 0.02883 |
| NCR+ILC3;A;CL | Absent |
Significance: 0.0201879 |
| ILC2_Nippo;A;CL | Absent |
Significance: 0.033467199999999996 |
| 416B_MP;CD34+;D;GEO | Absent |
Significance: 0.023221900000000004 |
| NCR+ILC3+IL23;A;CL | Absent |
Significance: 0.027512400000000003 |
| WHL_BRN;8wks;D;GEO | Absent |
Significance: 0.036434100000000004 |
| aB_LPS+IL4+Oligomycin_24h;A;CL | Absent |
Significance: 0.0450632 |
| FBRBLST;8wks;D;En | Absent |
Significance: 0.026382299999999997 |
| wCH12_drug;A;CL | Absent |
Significance: 0.0226155 |
| 3T3-L1;FBRBLST;D;En | Absent |
Significance: 0.0296493 |
| Mammary_GLND;A;GEO | Absent |
Significance: 0.0202674 |
| ILC1;A;GEO | Absent |
Significance: 0.0498513 |
| Muller;P12;D;En | Absent |
Significance: 0.0302837 |
| WHL_BRN;8wks;D;En | Absent |
Significance: 0.043396699999999996 |