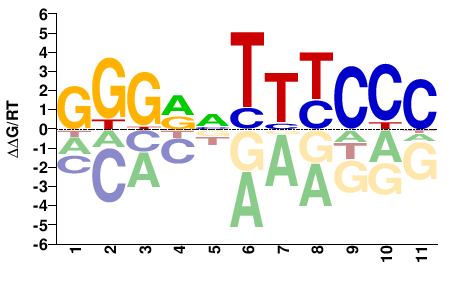
# Parameters
data_name = "Nfkb1"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:23:01,575 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Nfkb1: #46
| Cell type | Modisco | Homer |
|---|---|---|
| Th17;CD4+;A;CL |
Significance: 1.31019e-05 |
Significance: 4.09577e-05 |
| Spleen;8wks;D;En |
Significance: 1.52792e-05 |
Significance: 0.000478346 |
| B1_B;A;CL |
Significance: 0.000122402 |
Significance: 0.00151058 |
| aB_LPS;A;CL |
Significance: 9.32012e-05 |
Significance: 0.021300799999999998 |
| aB_LPS+IL21;A;CL |
Significance: 1.63107e-05 |
Significance: 0.00230666 |
| aB_LSP+a_d_dextran;A;CL |
Significance: 1.24335e-06 |
Significance: 0.00397416 |
| MCRPHG_BMD.LPD_Stmltd;A;GEO |
Significance: 2.38753e-05 |
Significance: 0.000217961 |
| DC_LPS;6h;A;GEO |
Significance: 1.01248e-06 |
Significance: 1.76739e-06 |
| DC_LPS;2h;A;GEO |
Significance: 1.18094e-06 |
Significance: 0.000256987 |
| CH12.LX;D;En |
Significance: 1.8891e-05 |
Significance: 1.02035e-05 |
| 3T3-L1;D;En |
Significance: 7.31686e-06 |
Significance: 0.0277942 |
| Tfh;A;CL |
Significance: 8.100119999999999e-05 |
Significance: 0.00975195 |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 4.76143e-07 |
Significance: 0.014602 |
| SKN_EPTHL;A;GEO |
Significance: 4.41419e-06 |
Significance: 0.0314457 |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL |
Significance: 7.02118e-05 |
Significance: 0.000341944 |
| DC_BMD;A;CL |
Significance: 1.67981e-05 |
Significance: 1.24719e-06 |
| ThMS_MEC;Aire++;A;GEO |
Significance: 0.000546505 |
Significance: 0.0021971 |
| Mammary_BptfKO;A;GEO |
Significance: 9.328260000000001e-09 |
Significance: 0.034128399999999996 |
| DC_LPS;4h;A;En |
Significance: 1.98397e-07 |
Significance: 0.00020852400000000002 |
| DC;A;En |
Significance: 1.5093e-07 |
Significance: 0.00057331 |
| aB;CD180;A;CL |
Significance: 0.00018047400000000002 |
Significance: 8.19549e-05 |
| gamma_delta_TCR_MCR;A;CL |
Significance: 1.1853900000000002e-05 |
Significance: 0.0008853089999999999 |
| apCD4Eff;D;GEO |
Significance: 3.35488e-05 |
Significance: 0.00248026 |
| ILC2precrsr;A;CL |
Significance: 5.38546e-05 |
Significance: 0.00171803 |
| NCR+ILC3;A;CL |
Significance: 5.9645e-06 |
Significance: 9.80264e-05 |
| ILC2_Nippo;A;CL |
Significance: 3.84432e-08 |
Significance: 3.04988e-06 |
| NCR+ILC3+IL23;A;CL |
Significance: 7.766519999999999e-06 |
Significance: 0.00147782 |
| ILC2;A;CL |
Significance: 1.0998099999999999e-05 |
Significance: 0.000199334 |
| Th2_nippo;A;CL |
Significance: 8.50007e-07 |
Significance: 7.06943e-05 |
| aB_LPS+IL4_24h;A;CL |
Significance: 3.96681e-05 |
Significance: 0.011800299999999998 |
| aB_LPS+IL4+Oligomycin_24h;A;CL |
Significance: 8.528e-05 |
Significance: 0.0006896119999999999 |
| CH12_QNTPLE;A;CL |
Significance: 5.09777e-06 |
Significance: 4.47776e-05 |
| wCH12;A;CL |
Significance: 3.67606e-05 |
Significance: 0.000157318 |
| CH12;med14;A;CL |
Significance: 2.9686000000000003e-06 |
Significance: 0.00234323 |
| wCH12_drug;A;CL |
Significance: 1.1410699999999999e-05 |
Significance: 3.18642e-05 |
| Mammary_GLND;A;GEO |
Significance: 4.75129e-06 |
Significance: 0.00455425 |
| ILC2+IL33+IL25;A;GEO |
Significance: 0.000259326 |
Significance: 3.38729e-05 |
| ILC1;A;GEO |
Significance: 3.06473e-05 |
Significance: 9.76732e-06 |
| ILC2;A;GEO |
Significance: 0.000137422 |
Significance: 0.000290866 |
| ILC3;A;GEO |
Significance: 0.000137086 |
Significance: 7.72326e-05 |
| ILC1_AbxGRMFree;A;GEO |
Significance: 8.538910000000001e-06 |
Significance: 0.00168223 |
| ILC2_AbxGRMFree;A;GEO |
Significance: 6.83607e-06 |
Significance: 0.024428599999999998 |
| ILC3_AbxGRMFree;A;GEO |
Significance: 2.1846200000000003e-08 |
Significance: 0.0137393 |
| abTreg;CD4+_CD25+;8wks;D;En |
Significance: 4.6482299999999994e-05 |
Significance: 0.0013731 |
| T_LCMV;CD8+;D33;A;GEO |
Significance: 9.47379e-05 |
Significance: 8.84633e-06 |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 8.713030000000001e-05 |
Significance: 0.0008387669999999999 |
Unique Cell types only TF-MoDISco finds Nfkb1: #54
| Cell type | Modisco | Homer |
|---|---|---|
| T_MLD;TCR_SV40-I_CD8+;D28;A;GEO |
Significance: 1.83106e-05 | Absent |
| memT;CD4+;A;CL |
Significance: 2.71769e-05 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D60;A;GEO |
Significance: 9.755030000000001e-06 | Absent |
| NK_SPLND;A;CL |
Significance: 5.9791000000000004e-05 | Absent |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 7.14052e-05 | Absent |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 7.58833e-07 | Absent |
| immatureB;A;CL |
Significance: 0.000798048 | Absent |
| matureB;A;CL |
Significance: 0.000321497 | Absent |
| MZ_B;A;CL |
Significance: 0.000336156 | Absent |
| memT_tmrD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 3.0001099999999997e-06 | Absent |
| GC_B;A;CL |
Significance: 0.00014942299999999998 | Absent |
| plasmaB;A;CL |
Significance: 0.00130381 | Absent |
| iNKT;A;GEO |
Significance: 0.00020743 | Absent |
| aB_LPS+IL4;A;CL |
Significance: 5.8070600000000006e-05 | Absent |
| aB;A;GEO |
Significance: 0.00016794 | Absent |
| prePLSM_B;A;GEO |
Significance: 5.4851999999999996e-05 | Absent |
| aB_CD40+IL4;A;CL |
Significance: 1.25949e-05 | Absent |
| B_SPLND;A;GEO |
Significance: 0.00123918 | Absent |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.000141424 | Absent |
| NTRPHL;A;CL |
Significance: 6.846559999999999e-07 | Absent |
| Tfh_LCMV;A;CL |
Significance: 1.77795e-05 | Absent |
| aNonTfh;A;CL |
Significance: 7.3535e-05 | Absent |
| aNonTfh_mut;A;CL |
Significance: 1.3625599999999999e-05 | Absent |
| Tfr;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 1.25468e-05 | Absent |
| naiveT;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.00032943699999999997 | Absent |
| naiveT_mut;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.000519336 | Absent |
| CD4+CD8intTCR+CD69+CD24+;B2defM;A;CL |
Significance: 0.00010062399999999999 | Absent |
| CD4+CD8-TCR+CD69+CD24+CD25-;B2defM;A;CL |
Significance: 0.00105915 | Absent |
| CD4+CD8-TCR+CD69-CD24-CD25-;B2defM;A;CL |
Significance: 0.00021202 | Absent |
| CD4+CD8+TCR+CD69+CD24+;MHC2defM;A;C |
Significance: 0.000872203 | Absent |
| CD4+CD8+TCR+CD69-CD24-;MHC2defM;A;CL |
Significance: 0.00155367 | Absent |
| CD4+CD8-TCR+CD25+Foxp3-;A;CL |
Significance: 0.00024234700000000002 | Absent |
| CD4+CD8-TCR+CD25-Foxp3+;A;CL |
Significance: 0.00025747599999999997 | Absent |
| Mammary;A;CL |
Significance: 4.67299e-07 | Absent |
| restingB;A;CL |
Significance: 5.528830000000001e-06 | Absent |
| gamma_delta_TCR;A;CL |
Significance: 1.2443299999999999e-06 | Absent |
| T_SPLND_blstCLTRD;CD4+;D;GEO |
Significance: 7.3754e-05 | Absent |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.000245065 | Absent |
| NK;IL2+IL12+;A;CL |
Significance: 0.00030378400000000003 | Absent |
| naiveT_SPLND;CD4+;D;GEO |
Significance: 2.18309e-06 | Absent |
| ILC1;A;CL |
Significance: 4.47714e-06 | Absent |
| FBRBLST;8wks;D;En |
Significance: 2.5213600000000002e-05 | Absent |
| mem_precrsrT_SPLND;CD8+;A;GEO |
Significance: 0.0002544 | Absent |
| NK_LVRD;CD19-CD3e-NKp46+DX+Trail-;A;GEO |
Significance: 0.00145224 | Absent |
| naiveT;CD8+;A;GEO |
Significance: 0.000142842 | Absent |
| T_LCMV;CD8+;D8;A;GEO |
Significance: 0.000108567 | Absent |
| TEff_SPLND;CD8+;A;GEO |
Significance: 7.13603e-05 | Absent |
| TExh_SPLND;CD8+;A;GEO |
Significance: 0.000218625 | Absent |
| TSLEC_SPLND;CD8+A;GEO |
Significance: 2.5050500000000004e-05 | Absent |
| naiveT;trnsgncTCR_SV40-I_CD8;A;GEO |
Significance: 0.00319285 | Absent |
| Teff;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 4.7766400000000004e-05 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D5;A;GEO |
Significance: 7.51162e-05 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.000198924 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D21;A;GEO |
Significance: 1.3177000000000001e-05 | Absent |
Unique Cell types only Homer finds Nfkb1: #8
| Cell type | Modisco | Homer |
|---|---|---|
| NPC;mE6;A;GEO | Absent |
Significance: 0.05460790000000001 |
| apThEff;CD4+;8wks;D;En. | Absent |
Significance: 0.00650009 |
| Treg;CD4+_CD25+;D;GEO | Absent |
Significance: 2.653e-05 |
| apTreg;8wk;D;En | Absent |
Significance: 0.0009283910000000001 |
| apTreg;D;GEO | Absent |
Significance: 8.70161e-05 |
| immatureB_SPLND;CD43-_CD11b-;D;GEO | Absent |
Significance: 0.000112727 |
| immatureB_SPLND;CD43-_CD11b-;D;En | Absent |
Significance: 0.0006308780000000001 |
| Tfh_mut;A;CL | Absent |
Significance: 4.76315e-05 |