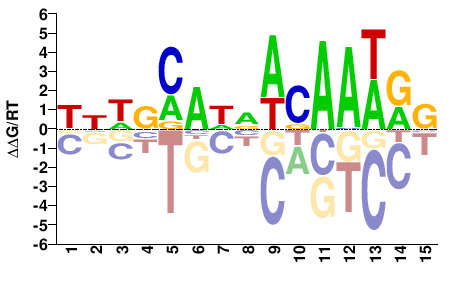
# Parameters
data_name = "Sox2"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:35:34,277 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Sox2: #36
| Cell type | Modisco | Homer |
|---|---|---|
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.0234541 |
Significance: 0.0226489 |
| ENDRM_ESD;A;GEO |
Significance: 0.00270511 |
Significance: 0.00011707 |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0171112 |
Significance: 2.00527e-05 |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.0226707 |
Significance: 9.606939999999999e-06 |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0215643 |
Significance: 0.00051925 |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.023253700000000002 |
Significance: 0.000534016 |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.0169918 |
Significance: 0.00016900400000000002 |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.0224196 |
Significance: 0.000868434 |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0197893 |
Significance: 0.000147302 |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.0157681 |
Significance: 0.00017623400000000002 |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.0195718 |
Significance: 0.0439794 |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.028275099999999997 |
Significance: 0.00103349 |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.0314126 |
Significance: 0.0008270160000000001 |
| NPC;F129;A;GEO |
Significance: 0.000857468 |
Significance: 0.0130589 |
| NPC;Ch13;A;GEO |
Significance: 0.0422903 |
Significance: 0.030806599999999996 |
| NPC;mE6;A;GEO |
Significance: 0.018194400000000003 |
Significance: 0.017694 |
| ES;E14;A;CL |
Significance: 4.71061e-06 |
Significance: 5.469740000000001e-06 |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.00361921 |
Significance: 2.36504e-05 |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.0110654 |
Significance: 0.041261400000000004 |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 2.1859e-06 |
Significance: 1.1041700000000001e-05 |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.000120864 |
Significance: 4.74289e-06 |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.0107973 |
Significance: 0.0414811 |
| ES;ZhBTc4;2;D;GEO |
Significance: 1.2827200000000001e-06 |
Significance: 1.40381e-07 |
| ES;D0;D;GEO |
Significance: 0.000162212 |
Significance: 1.29711e-06 |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 9.25364e-07 |
Significance: 7.87615e-08 |
| ES;E14;D;En |
Significance: 5.23418e-06 |
Significance: 5.102850000000001e-06 |
| ES;C57BL_6J;R1;D;En |
Significance: 3.52923e-06 |
Significance: 1.5776e-05 |
| ES;J1;A;CL |
Significance: 0.0174405 |
Significance: 2.0149900000000003e-06 |
| ES;E14;A;GEO |
Significance: 3.71786e-06 |
Significance: 2.28299e-06 |
| ES;F1216;A;GEO |
Significance: 2.8354400000000002e-06 |
Significance: 9.77417e-06 |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 3.2126799999999996e-06 |
Significance: 5.56194e-07 |
| ES;v6.5;A;GEO |
Significance: 3.7221499999999996e-06 |
Significance: 3.73906e-06 |
| wES;RTN;E11.5;D;En |
Significance: 0.024859 |
Significance: 0.0545135 |
| Muller;P12;D;En |
Significance: 0.00022557599999999998 |
Significance: 0.0185599 |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.026256900000000003 |
Significance: 0.00963551 |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.0403995 |
Significance: 0.00350816 |
Unique Cell types only TF-MoDISco finds Sox2: #77
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En |
Significance: 0.00732965 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D35;A;GEO |
Significance: 0.0561173 | Absent |
| wES;limb;E11.5;D;En |
Significance: 0.00273375 | Absent |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.00308911 | Absent |
| NK_SPLND;A;CL |
Significance: 0.05250219999999999 | Absent |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.00239602 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.00157764 | Absent |
| memT_tmrD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.0571399 | Absent |
| wES;FaceP;E11.5;D;En |
Significance: 0.00346663 | Absent |
| STMCH;P0;D;En |
Significance: 0.0344866 | Absent |
| B1_B;A;CL |
Significance: 0.0552174 | Absent |
| prePLSM_B;A;GEO |
Significance: 0.0532375 | Absent |
| SKLTL_Muscle;8wks;D;En |
Significance: 0.059509000000000006 | Absent |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.054653099999999996 | Absent |
| aB_LPS+IL4;A;CL |
Significance: 0.0448765 | Absent |
| Erythroid;Ter119+;A;GEO |
Significance: 0.0415177 | Absent |
| aB_CD40+IL4;A;CL |
Significance: 0.0524281 | Absent |
| Mast;A;CL |
Significance: 0.05917769999999999 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.05902090000000001 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.0321861 | Absent |
| naiveT_mut;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.0437105 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.00758625 | Absent |
| CD4+CD8-TCR+CD25+Foxp3-;A;CL |
Significance: 0.00515387 | Absent |
| CD4+CD8-TCR+CD25-Foxp3+;A;CL |
Significance: 0.042604500000000003 | Absent |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL |
Significance: 0.05023830000000001 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.020575299999999998 | Absent |
| MCRPHG;A;CL |
Significance: 0.0495025 | Absent |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 0.0360675 | Absent |
| NPC;G4;A;GEO |
Significance: 0.0007031510000000001 | Absent |
| DP3a;A;CL |
Significance: 0.053897400000000005 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.00732885 | Absent |
| eCortical_NRN;A;GEO |
Significance: 0.0186348 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.0575729 | Absent |
| ES;MEF;A;CL |
Significance: 0.0313697 | Absent |
| Muscle;A;CL |
Significance: 0.0469954 | Absent |
| Mammary;A;CL |
Significance: 6.35245e-05 | Absent |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 3.86537e-05 | Absent |
| Mammary_BptfKO;A;GEO |
Significance: 0.000395534 | Absent |
| placenta;A;CL |
Significance: 0.058337400000000005 | Absent |
| SPNL_CHRD;A;CL |
Significance: 7.89077e-05 | Absent |
| PNCRS;A;CL |
Significance: 0.0573595 | Absent |
| eye-NRL_RTN;A;CL |
Significance: 0.040487199999999994 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 1.69913e-05 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.0533019 | Absent |
| BRN;FRNTL_CRTX;A;CL |
Significance: 0.0408574 | Absent |
| BRN;PTMN;A;CL |
Significance: 0.000322571 | Absent |
| BRN;MDL_OBLNGT;A;CL |
Significance: 0.0007888410000000001 | Absent |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.00043694400000000004 | Absent |
| BRN;CRBLM;A;CL |
Significance: 0.045507 | Absent |
| aB;CD180;A;CL |
Significance: 0.059377099999999995 | Absent |
| LNG_TRM_HSC;A;CL |
Significance: 0.040232199999999996 | Absent |
| 3134_mammaryADNCRCNM;D;GE |
Significance: 0.0318192 | Absent |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.0597121 | Absent |
| ILC2precrsr;A;CL |
Significance: 0.0554679 | Absent |
| ILC2_Nippo;A;CL |
Significance: 0.0323262 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.00032690400000000004 | Absent |
| NCR+ILC3+IL23;A;CL |
Significance: 0.053191300000000004 | Absent |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.00142796 | Absent |
| FBRBLST;8wks;D;En |
Significance: 0.000536021 | Absent |
| KDNY;8wks;D;En |
Significance: 0.0329246 | Absent |
| wES;LNG;E14.5;D;En |
Significance: 0.0589842 | Absent |
| Mammary_GLND;A;GEO |
Significance: 0.00155503 | Absent |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.0187312 | Absent |
| Adipose;D;En |
Significance: 0.000614319 | Absent |
| RTN;D1;D;En |
Significance: 0.056594000000000005 | Absent |
| ILC3;A;GEO |
Significance: 0.05390219999999999 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 7.42861e-05 | Absent |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.042438699999999996 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.0397341 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.0267673 | Absent |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.0255255 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.0374239 | Absent |
| cntrl_memT;TCR_SV40-I_CD8+;A;GEO |
Significance: 0.0584861 | Absent |
| Md_BRN;D;En |
Significance: 0.0370744 | Absent |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.027162000000000002 | Absent |
| WHL_BRN;8wks;D;En |
Significance: 0.0471223 | Absent |
| wES;Telenchephalon;D;En |
Significance: 0.035276800000000004 | Absent |
Unique Cell types only Homer finds Sox2: #1
| Cell type | Modisco | Homer |
|---|---|---|
| aB;A;GEO | Absent |
Significance: 0.0580425 |