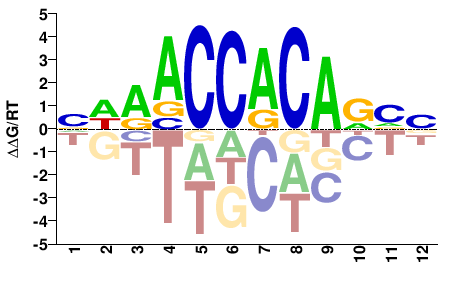
# Parameters
data_name = "Runx3"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:33:06,674 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Runx3: #59
| Cell type | Modisco | Homer |
|---|---|---|
| MPP;A;CL |
Significance: 0.00513599 |
Significance: 0.015511600000000002 |
| T_MLD;TCR_SV40-I_CD8+;D35;A;GEO |
Significance: 0.000130129 |
Significance: 0.000150707 |
| T_MLD;TCR_SV40-I_CD8+;D28;A;GEO |
Significance: 1.99712e-05 |
Significance: 8.0106e-08 |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.0005286140000000001 |
Significance: 0.0225209 |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.000181867 |
Significance: 0.0076295 |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D35;A;GEO |
Significance: 0.000637485 |
Significance: 0.0136685 |
| memT_tmrD;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.00023993 |
Significance: 0.000637684 |
| iNKT;A;GEO |
Significance: 0.00076953 |
Significance: 0.0110081 |
| Spleen;8wks;D;En |
Significance: 0.00221826 |
Significance: 0.00623105 |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.00016655900000000002 |
Significance: 0.00880667 |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.05837180000000001 |
Significance: 0.009142 |
| B;B220_CD43;A;GEO |
Significance: 0.00119188 |
Significance: 0.0158796 |
| aB_LPS+IL4;A;CL |
Significance: 0.00114286 |
Significance: 0.0306272 |
| MCRPHG_BMD.LPD_Stmltd;A;GEO |
Significance: 2.92076e-06 |
Significance: 0.00215339 |
| aB_LSP+a_d_dextran;A;CL |
Significance: 0.00151695 |
Significance: 0.0555274 |
| Mast;A;CL |
Significance: 1.14882e-06 |
Significance: 0.00020978599999999997 |
| BSPHL;A;CL |
Significance: 2.10611e-05 |
Significance: 0.00543236 |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 1.72919e-07 |
Significance: 0.0575149 |
| DC_LPS;2h;A;GEO |
Significance: 0.00035077 |
Significance: 0.00400777 |
| DC_pBMD;A;GEO |
Significance: 0.000842223 |
Significance: 0.010388799999999998 |
| 3T3-L1;D;En |
Significance: 0.00739133 |
Significance: 0.00116278 |
| naiveT;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.0101825 |
Significance: 0.0517783 |
| CD4+CD8intTCR+CD69+CD24+;B2defM;A;CL |
Significance: 0.00295921 |
Significance: 0.0501111 |
| CD4+CD8+TCR+CD69+CD24+;MHC2defM;A;C |
Significance: 0.00759957 |
Significance: 0.0111201 |
| CD4+CD8-TCR+CD25+Foxp3-;A;CL |
Significance: 0.011014399999999999 |
Significance: 0.00531986 |
| CD4+CD8-TCR+CD25-Foxp3+;A;CL |
Significance: 0.00999682 |
Significance: 0.0503539 |
| DC_BMD;A;CL |
Significance: 0.000210278 |
Significance: 1.59383e-05 |
| DP3a;A;CL |
Significance: 0.000249468 |
Significance: 0.0196779 |
| ES;MEF;A;CL |
Significance: 0.00046715900000000003 |
Significance: 0.0033189 |
| pCMP;A;CL |
Significance: 0.000455528 |
Significance: 0.011966899999999999 |
| DC_LPS;4h;A;En |
Significance: 3.73713e-05 |
Significance: 0.00911134 |
| pGMP;A;En |
Significance: 0.00719341 |
Significance: 0.011855500000000001 |
| DC;A;En |
Significance: 3.04994e-05 |
Significance: 0.000785468 |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.010808499999999999 |
Significance: 0.00115406 |
| LNG_TRM_HSC;A;CL |
Significance: 0.00172691 |
Significance: 0.045872699999999995 |
| 3134_mammaryADNCRCNM;D;GE |
Significance: 0.000542142 |
Significance: 0.0018534000000000003 |
| naiveT_SPLND;CD4+;D;GEO |
Significance: 0.00158397 |
Significance: 0.047712199999999996 |
| NK;IL2+IL12+;A;CL |
Significance: 2.1709699999999996e-05 |
Significance: 8.613719999999999e-05 |
| apCD4Eff;D;GEO |
Significance: 0.00190161 |
Significance: 0.0224315 |
| NCR+ILC3;A;CL |
Significance: 0.00017865400000000001 |
Significance: 0.0195542 |
| 416B_MP;CD34+;D;GEO |
Significance: 0.000250025 |
Significance: 0.00899539 |
| NCR+ILC3+IL23;A;CL |
Significance: 0.00059672 |
Significance: 0.014631899999999998 |
| CH12_QNTPLE;A;CL |
Significance: 6.992539999999999e-05 |
Significance: 0.0040654 |
| wCH12;A;CL |
Significance: 0.00151686 |
Significance: 0.00378261 |
| FBRBLST;8wks;D;En |
Significance: 0.000750838 |
Significance: 0.000542354 |
| 3T3-L1;FBRBLST;D;En |
Significance: 0.0124114 |
Significance: 0.00770363 |
| CLP;A;CL |
Significance: 0.00229089 |
Significance: 0.00516567 |
| mem_precrsrT_SPLND;CD8+;A;GEO |
Significance: 0.000150496 |
Significance: 0.00591601 |
| ILC1;A;GEO |
Significance: 7.86243e-05 |
Significance: 0.00183818 |
| ILC3;A;GEO |
Significance: 2.7351599999999997e-05 |
Significance: 0.00301741 |
| ILC1_AbxGRMFree;A;GEO |
Significance: 0.00122166 |
Significance: 0.0407651 |
| ILC3_AbxGRMFree;A;GEO |
Significance: 4.04291e-06 |
Significance: 0.00172431 |
| T_LCMV;CD8+;D8;A;GEO |
Significance: 0.00159404 |
Significance: 0.00020380099999999998 |
| Muller;P12;D;En |
Significance: 9.497869999999999e-06 |
Significance: 7.52721e-05 |
| TExh_SPLND;CD8+;A;GEO |
Significance: 0.00056394 |
Significance: 0.00264975 |
| TSLEC_SPLND;CD8+A;GEO |
Significance: 0.00012432 |
Significance: 0.00111942 |
| cntrl_memT;TCR_SV40-I_CD8+;A;GEO |
Significance: 0.00103099 |
Significance: 0.023668099999999997 |
| T_MLD;TCR_SV40-I_CD8+;D5;A;GEO |
Significance: 0.0008552039999999999 |
Significance: 0.031104400000000004 |
| T_MLD;TCR_SV40-I_CD8+;D21;A;GEO |
Significance: 0.000652767 |
Significance: 0.000371621 |
Unique Cell types only TF-MoDISco finds Runx3: #72
| Cell type | Modisco | Homer |
|---|---|---|
| naiveCD8;A;CL |
Significance: 0.000667658 | Absent |
| memT;CD4+;A;CL |
Significance: 0.00292645 | Absent |
| Th17;CD4+;A;CL |
Significance: 0.00199533 | Absent |
| NK_SPLND;A;CL |
Significance: 0.00011180299999999999 | Absent |
| proB;A;CL |
Significance: 0.000320952 | Absent |
| preB;A;CL |
Significance: 0.00152721 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D60;A;GEO |
Significance: 0.000603027 | Absent |
| matureB;A;CL |
Significance: 0.00282441 | Absent |
| MZ_B;A;CL |
Significance: 0.047051800000000005 | Absent |
| B1_B;A;CL |
Significance: 0.00466792 | Absent |
| memT_tmrD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.000400995 | Absent |
| plasmaB;A;CL |
Significance: 0.00286155 | Absent |
| aB;A;GEO |
Significance: 0.00442324 | Absent |
| preB_BMD;A;GEO |
Significance: 0.00022795200000000002 | Absent |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.00178675 | Absent |
| AML;D;En |
Significance: 4.04011e-05 | Absent |
| AML;AE9a;A;GEO |
Significance: 0.00039740800000000003 | Absent |
| DC_LPS;6h;A;GEO |
Significance: 0.000759903 | Absent |
| MEL;D;En |
Significance: 0.00792383 | Absent |
| Tfh_LCMV;A;CL |
Significance: 0.0102417 | Absent |
| Tfh;A;CL |
Significance: 0.00545241 | Absent |
| aNonTfh;A;CL |
Significance: 0.00157796 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.00028462400000000003 | Absent |
| Tfr;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.00139056 | Absent |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.00276993 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.00109678 | Absent |
| naiveT_mut;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.000282185 | Absent |
| largeDPprecrsr;CD4+CD8+CD69-;A;CL |
Significance: 0.010580899999999999 | Absent |
| smallDPprecrsor;CD4+CD8+CD69-;A;CL |
Significance: 7.68843e-05 | Absent |
| CD4+CD8-TCR+CD69+CD24+CD25-;B2defM;A;CL |
Significance: 0.00457167 | Absent |
| CD4+CD8-TCR+CD69-CD24-CD25-;B2defM;A;CL |
Significance: 0.00365566 | Absent |
| CD4+CD8intTCR+CD69+CD24+;MHC2defM;A;CL |
Significance: 0.00406148 | Absent |
| CD4+CD8+TCR+CD69-CD24-;MHC2defM;A;CL |
Significance: 0.00319809 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.00129349 | Absent |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL |
Significance: 0.00409875 | Absent |
| MCRPHG;A;CL |
Significance: 0.0225185 | Absent |
| DP;A;CL |
Significance: 0.00149436 | Absent |
| DP3b;A;CL |
Significance: 0.00155753 | Absent |
| ISP;A;CL |
Significance: 0.00319297 | Absent |
| ThMS_MEC;Aire++;A;GEO |
Significance: 0.00633172 | Absent |
| Mammary_BptfKO;A;GEO |
Significance: 0.00297967 | Absent |
| pMeg;A;En |
Significance: 0.00056139 | Absent |
| MegP;A;En |
Significance: 0.00297268 | Absent |
| aB;CD180;A;CL |
Significance: 0.00037276400000000004 | Absent |
| SHRT_TRM_HSC;A;CL |
Significance: 0.000720837 | Absent |
| gamma_delta_TCR;A;CL |
Significance: 0.000985073 | Absent |
| gamma_delta_TCR_MCR;A;CL |
Significance: 0.00717786 | Absent |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.000633025 | Absent |
| ILC2precrsr;A;CL |
Significance: 0.0245803 | Absent |
| ILC2_Nippo;A;CL |
Significance: 0.00917839 | Absent |
| ILC1;A;CL |
Significance: 0.000326571 | Absent |
| ILC2;A;CL |
Significance: 0.00576287 | Absent |
| Th2_nippo;A;CL |
Significance: 0.00620637 | Absent |
| aB_LPS+IL4_24h;A;CL |
Significance: 0.00435057 | Absent |
| aB_LPS+IL4+Oligomycin_24h;A;CL |
Significance: 2.35917e-05 | Absent |
| NIH3T3_FBROBLST;D;En |
Significance: 0.0123191 | Absent |
| wCH12_drug;A;CL |
Significance: 8.13084e-05 | Absent |
| NK_LVRD;CD19-CD3e-NKp46+DX+Trail-;A;GEO |
Significance: 3.06175e-07 | Absent |
| Mammary_GLND;A;GEO |
Significance: 0.000136813 | Absent |
| ILC2+IL33+IL25;A;GEO |
Significance: 0.000721026 | Absent |
| ILC2;A;GEO |
Significance: 0.00219423 | Absent |
| abTreg;CD4+_CD25+;8wks;D;En |
Significance: 0.00268804 | Absent |
| ILC2_AbxGRMFree;A;GEO |
Significance: 0.00271263 | Absent |
| naiveT;CD8+;A;GEO |
Significance: 0.000298815 | Absent |
| 416B_MP;CD34+;D;En |
Significance: 0.000247435 | Absent |
| T_LCMV;CD8+;D33;A;GEO |
Significance: 0.000457822 | Absent |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.00030267200000000003 | Absent |
| TEff_SPLND;CD8+;A;GEO |
Significance: 0.00159792 | Absent |
| naiveT;trnsgncTCR_SV40-I_CD8;A;GEO |
Significance: 0.000359565 | Absent |
| Teff;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.000393291 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.000692479 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 6.0274e-05 | Absent |
Unique Cell types only Homer finds Runx3: #5
| Cell type | Modisco | Homer |
|---|---|---|
| prePLSM_B;A;GEO | Absent |
Significance: 0.044464800000000006 |
| B_SPLND;A;GEO | Absent |
Significance: 0.0381948 |
| apTreg;D;GEO | Absent |
Significance: 0.0337742 |
| MEL_Znc_di-cl;24h;D;En | Absent |
Significance: 0.00248041 |
| Tfh_mut;A;CL | Absent |
Significance: 0.0352885 |