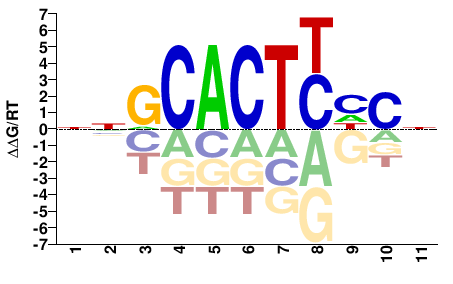
# Parameters
data_name = "Zscan10"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:54:29,155 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Zscan10: #5
| Cell type | Modisco | Homer |
|---|---|---|
| CD4+CD8-TCR+CD69+CD24+CD25-;B2defM;A;CL |
Significance: 0.0364346 |
Significance: 0.0526249 |
| restingB;A;CL |
Significance: 0.0087715 |
Significance: 0.019677700000000003 |
| B;B220_CD43;A;GEO |
Significance: 0.014393000000000001 |
Significance: 0.0427776 |
| gamma_delta_TCR_MCR;A;CL |
Significance: 0.0148665 |
Significance: 0.0567446 |
| CH12.LX;D;En |
Significance: 0.027514599999999997 |
Significance: 0.033253899999999996 |
Unique Cell types only TF-MoDISco finds Zscan10: #89
| Cell type | Modisco | Homer |
|---|---|---|
| naiveCD8;A;CL |
Significance: 0.030114799999999997 | Absent |
| memT;CD4+;A;CL |
Significance: 0.0357574 | Absent |
| Th17;CD4+;A;CL |
Significance: 0.028747 | Absent |
| NK_SPLND;A;CL |
Significance: 0.039636300000000006 | Absent |
| proB;A;CL |
Significance: 0.0194126 | Absent |
| preB;A;CL |
Significance: 0.0104969 | Absent |
| immatureB;A;CL |
Significance: 0.00820657 | Absent |
| matureB;A;CL |
Significance: 0.0079541 | Absent |
| MZ_B;A;CL |
Significance: 0.011446 | Absent |
| B1_B;A;CL |
Significance: 0.0102596 | Absent |
| GC_B;A;CL |
Significance: 0.00714633 | Absent |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.05526709999999999 | Absent |
| aB_LPS;A;CL |
Significance: 0.00879301 | Absent |
| aB_LPS+IL4;A;CL |
Significance: 0.036408800000000005 | Absent |
| aB_LPS+IL21;A;CL |
Significance: 0.0145671 | Absent |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.051449 | Absent |
| aB_CD40+IL4;A;CL |
Significance: 0.013276900000000001 | Absent |
| A20;B-lymphoma;D;En |
Significance: 0.00630495 | Absent |
| AML;D;En |
Significance: 0.00722457 | Absent |
| aNonTfh;A;CL |
Significance: 0.05184400000000001 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.046228899999999996 | Absent |
| naiveT;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.044827 | Absent |
| largeDPprecrsr;CD4+CD8+CD69-;A;CL |
Significance: 0.0409028 | Absent |
| CD4+CD8-TCR+CD69-CD24-CD25-;B2defM;A;CL |
Significance: 0.050425199999999996 | Absent |
| CD4+CD8+TCR+CD69-CD24-;MHC2defM;A;CL |
Significance: 0.0449268 | Absent |
| CD4+CD8-TCR+CD25+Foxp3-;A;CL |
Significance: 0.037148400000000005 | Absent |
| CD4+CD8-TCR+CD25-Foxp3+;A;CL |
Significance: 0.0425857 | Absent |
| MCRPHG;A;CL |
Significance: 0.00861557 | Absent |
| ISP;A;CL |
Significance: 0.03171090000000001 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.049980300000000005 | Absent |
| Heart;D;En |
Significance: 0.0541428 | Absent |
| Adipose;A;CL |
Significance: 0.026025599999999996 | Absent |
| gamma_delta_TCR;A;CL |
Significance: 0.0506515 | Absent |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.0310666 | Absent |
| ILC2precrsr;A;CL |
Significance: 0.0371856 | Absent |
| ILC2_Nippo;A;CL |
Significance: 0.0387056 | Absent |
| aB_LPS+IL4_24h;A;CL |
Significance: 0.0213992 | Absent |
| CH12_QNTPLE;A;CL |
Significance: 0.00674395 | Absent |
| wCH12;A;CL |
Significance: 0.05219209999999999 | Absent |
| wCH12_drug;A;CL |
Significance: 0.0180162 | Absent |
| CLP;A;CL |
Significance: 0.00805431 | Absent |
| mem_precrsrT_SPLND;CD8+;A;GEO |
Significance: 0.041053 | Absent |
| ES;J1;A;CL |
Significance: 0.0194888 | Absent |
| ES;F1216;A;GEO |
Significance: 0.0537047 | Absent |
| ILC2+IL33+IL25;A;GEO |
Significance: 0.0316886 | Absent |
| ILC2;A;GEO |
Significance: 0.037228500000000005 | Absent |
| ILC3;A;GEO |
Significance: 0.03490019999999999 | Absent |
| ILC1_AbxGRMFree;A;GEO |
Significance: 0.055568200000000005 | Absent |
| naiveT;CD8+;A;GEO |
Significance: 0.0416394 | Absent |
| T_LCMV;CD8+;D33;A;GEO |
Significance: 0.0458384 | Absent |
| TEff_SPLND;CD8+;A;GEO |
Significance: 0.055913199999999996 | Absent |
| T_MLD;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.0377616 | Absent |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.0293402 | Absent |
| aB;A;GEO |
Significance: 0.015447799999999998 | Absent |
| B_SPLND;A;GEO |
Significance: 0.014827799999999999 | Absent |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.00755072 | Absent |
| MCRPHG_BMD.LPD_Stmltd;A;GEO |
Significance: 0.0290575 | Absent |
| AML;AE9a;A;GEO |
Significance: 0.05205219999999999 | Absent |
| DC_LPS;6h;A;GEO |
Significance: 0.0266454 | Absent |
| DC_pBMD;A;GEO |
Significance: 0.0117456 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.05536749999999999 | Absent |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.059142099999999996 | Absent |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.05837230000000001 | Absent |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0545797 | Absent |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.045787699999999994 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.052798000000000005 | Absent |
| iPV_NRN;A;GEO |
Significance: 0.0450212 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.0585534 | Absent |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.046079 | Absent |
| pMeg;A;En |
Significance: 0.00637353 | Absent |
| MegP;A;En |
Significance: 0.010341 | Absent |
| pCMP;A;CL |
Significance: 0.00890798 | Absent |
| DC_LPS;4h;A;En |
Significance: 0.0375514 | Absent |
| pGMP;A;En |
Significance: 0.00735863 | Absent |
| DC;A;En |
Significance: 0.0212953 | Absent |
| T_SPLND_blstCLTRD;CD4+;D;GEO |
Significance: 0.038458 | Absent |
| immatureB_SPLND;CD43-_CD11b-;D;GEO |
Significance: 0.00893613 | Absent |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.05785030000000001 | Absent |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 0.0484376 | Absent |
| ES;C57BL_6J;R1;D;En |
Significance: 0.0548703 | Absent |
| FBRBLST;8wks;D;En |
Significance: 0.0487149 | Absent |
| KDNY;P0;D;En |
Significance: 0.0542715 | Absent |
| LNG;P0;D;En |
Significance: 0.0374815 | Absent |
| wES;LNG;E14.5;D;En |
Significance: 0.0533239 | Absent |
| Heart;P0;D;En |
Significance: 0.05183340000000001 | Absent |
| immatureB_SPLND;CD43-_CD11b-;D;En |
Significance: 0.00838074 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 0.0564367 | Absent |
| Muller;P12;D;En |
Significance: 0.05816 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.0532288 | Absent |
Unique Cell types only Homer finds Zscan10: #5
| Cell type | Modisco | Homer |
|---|---|---|
| iNKT;A;GEO | Absent |
Significance: 0.0357056 |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL | Absent |
Significance: 0.027249400000000003 |
| DP;A;CL | Absent |
Significance: 0.051097199999999995 |
| T_LCMV;CD8+;D35;A;GEO | Absent |
Significance: 0.05755830000000001 |
| T_MLD;TCR_SV40-I_CD8+;D14;A;GEO | Absent |
Significance: 0.0365806 |