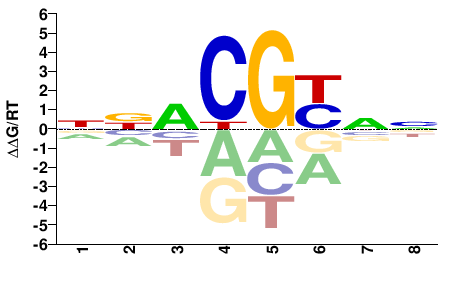
# Parameters
data_name = "Gmeb2"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:10:45,715 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Gmeb2: #14
| Cell type | Modisco | Homer |
|---|---|---|
| LVR;8wks;D;En |
Significance: 0.0434879 |
Significance: 0.0197238 |
| NTRPHL;A;GEO |
Significance: 0.0338513 |
Significance: 0.0123113 |
| LNG;8wks;D;En |
Significance: 0.0192247 |
Significance: 0.0157221 |
| LNG;P0;D;En |
Significance: 0.0310841 |
Significance: 0.0142012 |
| LVR;8wks;D;GEO |
Significance: 0.0370017 |
Significance: 0.0144733 |
| DC_BMD;A;CL |
Significance: 0.0385922 |
Significance: 0.050739599999999996 |
| LNG;8wks;D;GEO |
Significance: 0.0229082 |
Significance: 0.0241829 |
| CRBLM;D;En |
Significance: 0.0557163 |
Significance: 0.0200942 |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.0574461 |
Significance: 0.018889700000000002 |
| BSPHL;A;CL |
Significance: 0.0440386 |
Significance: 0.0299846 |
| DC_LPS;2h;A;GEO |
Significance: 0.0557865 |
Significance: 0.00286735 |
| NTRPHL;A;CL |
Significance: 0.042917500000000004 |
Significance: 0.031209300000000002 |
| Muscle;A;CL |
Significance: 0.05410269999999999 |
Significance: 0.0248543 |
| Mammary_BptfKO;A;GEO |
Significance: 0.0283493 |
Significance: 0.059072400000000004 |
Unique Cell types only TF-MoDISco finds Gmeb2: #68
| Cell type | Modisco | Homer |
|---|---|---|
| naiveCD8;A;CL |
Significance: 0.0129345 | Absent |
| memT;CD4+;A;CL |
Significance: 0.0514245 | Absent |
| Th17;CD4+;A;CL |
Significance: 0.0403318 | Absent |
| NK_SPLND;A;CL |
Significance: 0.036062000000000004 | Absent |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.0389394 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.0392664 | Absent |
| LRG_INTSTN;8wks;D;En |
Significance: 0.0229866 | Absent |
| matureB;A;CL |
Significance: 0.027485700000000002 | Absent |
| aB;A;GEO |
Significance: 0.0429834 | Absent |
| STMCH;P0;D;En |
Significance: 0.010344200000000001 | Absent |
| B_SPLND;A;GEO |
Significance: 0.0301666 | Absent |
| plasmaB;A;CL |
Significance: 0.0481891 | Absent |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.029870499999999998 | Absent |
| MEL;D;En |
Significance: 0.012852299999999999 | Absent |
| Mast;A;CL |
Significance: 0.0217942 | Absent |
| MEL_Znc_di-cl;24h;D;En |
Significance: 0.0149784 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.00796271 | Absent |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.0270947 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.049525599999999996 | Absent |
| Tfr;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.053036099999999996 | Absent |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.0416295 | Absent |
| CD4+CD8-TCR+CD69+CD24+CD25-;B2defM;A;CL |
Significance: 0.026270299999999996 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.054624599999999995 | Absent |
| SKN_EPTHL;A;GEO |
Significance: 0.05920159999999999 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.0200581 | Absent |
| NPC;F129;A;GEO |
Significance: 0.052209000000000005 | Absent |
| iPV_NRN;A;GEO |
Significance: 0.0314719 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.048179900000000005 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.0399327 | Absent |
| ES;MEF;A;CL |
Significance: 0.0296909 | Absent |
| ThMS_MEC;Aire++;A;GEO |
Significance: 0.017365099999999998 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.019371400000000004 | Absent |
| Melanotrope;A;GEO |
Significance: 0.00748328 | Absent |
| Heart;D;En |
Significance: 0.042269 | Absent |
| testis;A;CL |
Significance: 0.0410351 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.0520394 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.026158799999999996 | Absent |
| BRN;PTMN;A;CL |
Significance: 0.033556300000000004 | Absent |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.0573193 | Absent |
| BRN;CRBLM;A;CL |
Significance: 0.00928499 | Absent |
| restingB;A;CL |
Significance: 0.0447726 | Absent |
| LNG_TRM_HSC;A;CL |
Significance: 0.0105738 | Absent |
| gamma_delta_TCR;A;CL |
Significance: 0.0576668 | Absent |
| gamma_delta_TCR_MCR;A;CL |
Significance: 0.055774199999999996 | Absent |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.0232438 | Absent |
| NK;IL2+IL12+;A;CL |
Significance: 0.0559649 | Absent |
| naiveT_SPLND;CD4+;D;GEO |
Significance: 0.017042599999999998 | Absent |
| NCR+ILC3;A;CL |
Significance: 0.015386700000000001 | Absent |
| ILC2_Nippo;A;CL |
Significance: 0.029544499999999998 | Absent |
| ILC1;A;CL |
Significance: 0.0547183 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.0247603 | Absent |
| ILC2;A;CL |
Significance: 0.0338886 | Absent |
| KDNY;P0;D;En |
Significance: 0.00620382 | Absent |
| KDNY;8wks;D;En |
Significance: 0.0398546 | Absent |
| wES;LVR;E11.5;D.;En |
Significance: 0.00962002 | Absent |
| Heart;8wks;D;En |
Significance: 0.0118771 | Absent |
| Mammary_GLND;A;GEO |
Significance: 0.0502344 | Absent |
| Heart;P0;D;En |
Significance: 0.0291714 | Absent |
| ILC2+IL33+IL25;A;GEO |
Significance: 0.037723400000000004 | Absent |
| ILC1;A;GEO |
Significance: 0.057815599999999995 | Absent |
| ILC2;A;GEO |
Significance: 0.0237437 | Absent |
| ILC3;A;GEO |
Significance: 0.051410199999999996 | Absent |
| ILC1_AbxGRMFree;A;GEO |
Significance: 0.029677099999999994 | Absent |
| ILC3_AbxGRMFree;A;GEO |
Significance: 0.0455657 | Absent |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.0479419 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.023922299999999997 | Absent |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.0587004 | Absent |
| WHL_BRN;8wks;D;En |
Significance: 0.0245668 | Absent |
Unique Cell types only Homer finds Gmeb2: #20
| Cell type | Modisco | Homer |
|---|---|---|
| MegP;A;En | Absent |
Significance: 0.00635609 |
| DC_LPS;4h;A;En | Absent |
Significance: 0.019661900000000003 |
| pGMP;A;En | Absent |
Significance: 0.0181623 |
| DC;A;En | Absent |
Significance: 0.00604287 |
| SKLTL_Muscle;8wks;D;En | Absent |
Significance: 0.015788999999999997 |
| NIH3T3_FBROBLST;D;GEO | Absent |
Significance: 0.0546268 |
| Adipose;D;En | Absent |
Significance: 0.0163807 |
| AML;D;En | Absent |
Significance: 0.0225612 |
| NIH3T3_FBROBLST;D;En | Absent |
Significance: 0.0214715 |
| 416B_MP;CD34+;D;En | Absent |
Significance: 0.00772696 |
| DC_LPS;6h;A;GEO | Absent |
Significance: 0.00307113 |
| NIH3T3;D;En | Absent |
Significance: 0.00841401 |
| placenta;A;CL | Absent |
Significance: 0.028111900000000002 |
| 416B_MP;CD34+;D;GEO | Absent |
Significance: 0.00951435 |
| 3T3-L1;D;En | Absent |
Significance: 0.00680347 |
| Muller;P12;D;En | Absent |
Significance: 0.0298319 |
| Teff;TCR_SV40-I_CD8+;D7;A;GEO | Absent |
Significance: 0.045677499999999996 |
| Adrenal;A;CL | Absent |
Significance: 0.029389799999999997 |
| FBRBLST;8wks;D;En | Absent |
Significance: 0.0271411 |
| pMeg;A;En | Absent |
Significance: 0.00992602 |