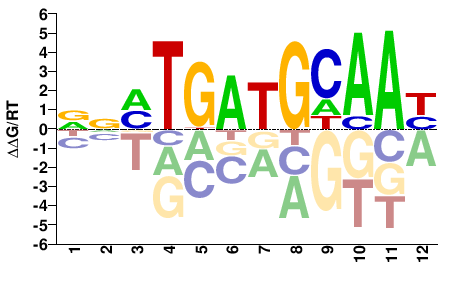
# Parameters
data_name = "Cebpg"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:00:11,295 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Cebpg: #45
| Cell type | Modisco | Homer |
|---|---|---|
| SKLTL_Muscle;8wks;D;En |
Significance: 0.00747995 |
Significance: 0.0336503 |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.00246917 |
Significance: 0.0048878 |
| AML;D;En |
Significance: 0.0095262 |
Significance: 0.00812466 |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.0148351 |
Significance: 0.0184035 |
| MCRPHG_BMD.LPD_Stmltd;A;GEO |
Significance: 0.00451324 |
Significance: 0.010151299999999999 |
| DC_LPS;6h;A;GEO |
Significance: 0.00620445 |
Significance: 0.0205457 |
| BSPHL;A;CL |
Significance: 0.00329677 |
Significance: 0.00019292900000000002 |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 0.00797096 |
Significance: 0.00208241 |
| DC_LPS;2h;A;GEO |
Significance: 0.01243 |
Significance: 0.017885400000000003 |
| NTRPHL;A;CL |
Significance: 0.0355206 |
Significance: 0.0339541 |
| DC_pBMD;A;GEO |
Significance: 0.011289200000000001 |
Significance: 0.0187065 |
| NIH3T3;D;En |
Significance: 0.00326175 |
Significance: 0.0598403 |
| 3T3-L1;D;En |
Significance: 0.0561846 |
Significance: 0.020192500000000002 |
| DC_BMD;A;CL |
Significance: 0.0474056 |
Significance: 0.00257087 |
| MCRPHG;A;CL |
Significance: 0.0170536 |
Significance: 0.0199293 |
| iPV_NRN;A;GEO |
Significance: 0.0529481 |
Significance: 0.045108999999999996 |
| Adrenal;A;CL |
Significance: 0.00745781 |
Significance: 0.05700830000000001 |
| Mammary_BptfKO;A;GEO |
Significance: 0.00333855 |
Significance: 0.00178108 |
| placenta;A;CL |
Significance: 0.0104241 |
Significance: 0.00987777 |
| pMeg;A;En |
Significance: 0.0045601999999999995 |
Significance: 0.00440051 |
| MegP;A;En |
Significance: 0.00618001 |
Significance: 0.00129252 |
| pCMP;A;CL |
Significance: 0.0203526 |
Significance: 0.00888967 |
| DC_LPS;4h;A;En |
Significance: 0.00212292 |
Significance: 0.0408323 |
| pGMP;A;En |
Significance: 0.00388044 |
Significance: 0.013391499999999999 |
| DC;A;En |
Significance: 0.00506571 |
Significance: 0.047443400000000004 |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.007327199999999999 |
Significance: 0.0009311710000000001 |
| LVR;8wks;D;GEO |
Significance: 0.00546605 |
Significance: 0.0170222 |
| LNG;8wks;D;GEO |
Significance: 0.00511745 |
Significance: 0.00103542 |
| 416B_MP;CD34+;D;GEO |
Significance: 0.00354701 |
Significance: 0.00115319 |
| NIH3T3_FBROBLST;D;En |
Significance: 0.00433116 |
Significance: 0.055908400000000004 |
| FBRBLST;8wks;D;En |
Significance: 0.00588184 |
Significance: 0.0335107 |
| 3T3-L1;FBRBLST;D;En |
Significance: 0.00278397 |
Significance: 1.39606e-06 |
| LVR;8wks;D;En |
Significance: 0.004285199999999999 |
Significance: 0.0289308 |
| NTRPHL;A;GEO |
Significance: 0.014772799999999997 |
Significance: 0.00184941 |
| wES;LVR;E11.5;D.;En |
Significance: 0.00511826 |
Significance: 0.0046799 |
| LVR;P0;D;En |
Significance: 0.00962292 |
Significance: 0.00324762 |
| LNG;8wks;D;En |
Significance: 0.00512746 |
Significance: 0.00764528 |
| LNG;P0;D;En |
Significance: 0.00372541 |
Significance: 0.0544285 |
| Mammary_GLND;A;GEO |
Significance: 0.00211786 |
Significance: 0.00509131 |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 0.00690075 |
Significance: 0.00084583 |
| Adipose;D;En |
Significance: 0.0374129 |
Significance: 0.00691067 |
| CRBLM;D;En |
Significance: 0.027162000000000002 |
Significance: 0.0531846 |
| 416B_MP;CD34+;D;En |
Significance: 0.00595661 |
Significance: 0.00999437 |
| Muller;P12;D;En |
Significance: 0.00374945 |
Significance: 0.041995099999999994 |
| WHL_BRN;8wks;D;En |
Significance: 0.0435431 |
Significance: 0.0321861 |
Unique Cell types only TF-MoDISco finds Cebpg: #53
| Cell type | Modisco | Homer |
|---|---|---|
| MPP;A;CL |
Significance: 0.00612123 | Absent |
| naiveCD8;A;CL |
Significance: 0.0581974 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.055344700000000004 | Absent |
| wES;FaceP;E11.5;D;En |
Significance: 0.05777519999999999 | Absent |
| LRG_INTSTN;8wks;D;En |
Significance: 0.0405837 | Absent |
| STMCH;P0;D;En |
Significance: 0.050194 | Absent |
| aB;A;GEO |
Significance: 0.059635400000000005 | Absent |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.059099 | Absent |
| CH12.LX;D;En |
Significance: 0.055195400000000006 | Absent |
| AML;AE9a;A;GEO |
Significance: 0.059478699999999995 | Absent |
| MEL;D;En |
Significance: 0.00964828 | Absent |
| MEL_Znc_di-cl;24h;D;En |
Significance: 0.0122949 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.0350175 | Absent |
| ESNPHL_SPLND_BlbCA;CL |
Significance: 0.00334253 | Absent |
| MN1;D;En |
Significance: 0.05255309999999999 | Absent |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.0271413 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.05250740000000001 | Absent |
| SKN_EPTHL;A;GEO |
Significance: 0.0224515 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.0326876 | Absent |
| NPC;F129;A;GEO |
Significance: 0.0440773 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.0433556 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.054127800000000004 | Absent |
| ES;MEF;A;CL |
Significance: 0.00190251 | Absent |
| ThMS_MEC;Aire++;A;GEO |
Significance: 0.030776400000000002 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.0468561 | Absent |
| LNG;A;CL |
Significance: 0.00485608 | Absent |
| Muscle;A;CL |
Significance: 0.04643869999999999 | Absent |
| Heart;D;En |
Significance: 0.019538 | Absent |
| Corticotrope;A;GEO |
Significance: 0.0248578 | Absent |
| Mammary;A;CL |
Significance: 0.00836772 | Absent |
| Melanotrope;A;GEO |
Significance: 0.0510587 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.041425 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.0507271 | Absent |
| BRN;FRNTL_CRTX;A;CL |
Significance: 0.037266900000000006 | Absent |
| BRN;PTMN;A;CL |
Significance: 0.059602999999999996 | Absent |
| BRN;CRBLM;A;CL |
Significance: 0.0381561 | Absent |
| LNG_TRM_HSC;A;CL |
Significance: 0.029692200000000002 | Absent |
| 3134_mammaryADNCRCNM;D;GE |
Significance: 0.0101934 | Absent |
| naiveT_SPLND;CD4+;D;GEO |
Significance: 0.0501904 | Absent |
| gamma_delta_TCR_M-S-;A;CL |
Significance: 0.0541345 | Absent |
| NCR+ILC3;A;CL |
Significance: 0.0557759 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.0591312 | Absent |
| wCH12_drug;A;CL |
Significance: 1.79815e-07 | Absent |
| KDNY;P0;D;En |
Significance: 0.030308699999999997 | Absent |
| KDNY;8wks;D;En |
Significance: 0.0329246 | Absent |
| wES;LNG;E14.5;D;En |
Significance: 0.0284022 | Absent |
| Heart;8wks;D;En |
Significance: 0.030303700000000003 | Absent |
| Heart;P0;D;En |
Significance: 0.05949490000000001 | Absent |
| ILC2;A;GEO |
Significance: 0.0435113 | Absent |
| ILC3;A;GEO |
Significance: 0.0358279 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.044789300000000004 | Absent |
| Md_BRN;D;En |
Significance: 0.051963499999999996 | Absent |
| wES;Telenchephalon;D;En |
Significance: 0.0490285 | Absent |
Unique Cell types only Homer finds Cebpg: #3
| Cell type | Modisco | Homer |
|---|---|---|
| restingB;A;CL | Absent |
Significance: 0.0587057 |
| ILC1_AbxGRMFree;A;GEO | Absent |
Significance: 0.0205273 |
| aB_LPS+IL21;A;CL | Absent |
Significance: 0.0564212 |