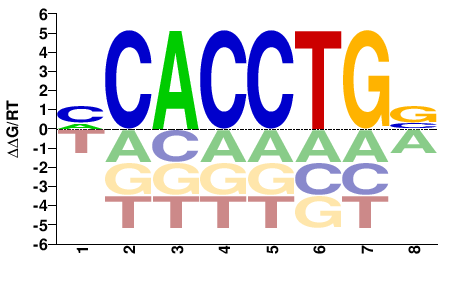
# Parameters
data_name = "Snai1"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:34:37,953 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Snai1: #12
| Cell type | Modisco | Homer |
|---|---|---|
| largeDPprecrsr;CD4+CD8+CD69-;A;CL |
Significance: 0.0106137 |
Significance: 0.0565764 |
| smallDPprecrsor;CD4+CD8+CD69-;A;CL |
Significance: 0.0140255 |
Significance: 0.042062300000000004 |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.0188207 |
Significance: 0.0570976 |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.05098130000000001 |
Significance: 0.0139918 |
| RTN;D1;D;En |
Significance: 0.0242874 |
Significance: 0.0480688 |
| NPC;G4;A;GEO |
Significance: 0.021516 |
Significance: 0.052487599999999995 |
| plasmaB;A;CL |
Significance: 0.00382078 |
Significance: 0.040282 |
| NPC;mE6;A;GEO |
Significance: 0.0422182 |
Significance: 0.05612140000000001 |
| DP;A;CL |
Significance: 0.013592600000000002 |
Significance: 0.0403263 |
| DP3b;A;CL |
Significance: 0.0170666 |
Significance: 0.0530831 |
| ISP;A;CL |
Significance: 0.0507753 |
Significance: 0.041265699999999995 |
| wES;MSDRM;E11.5;D;En |
Significance: 0.0320157 |
Significance: 0.00926239 |
Unique Cell types only TF-MoDISco finds Snai1: #42
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E11.5;D;En |
Significance: 0.0194102 | Absent |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.0279178 | Absent |
| proB;A;CL |
Significance: 0.0244199 | Absent |
| preB;A;CL |
Significance: 0.0148602 | Absent |
| preB_BMD;A;GEO |
Significance: 0.01395 | Absent |
| B;B220_CD43;A;GEO |
Significance: 0.019976400000000002 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.031775599999999994 | Absent |
| MN1;D;En |
Significance: 0.045321600000000004 | Absent |
| CN_PHT_RCPTR;A;GEO |
Significance: 0.057475 | Absent |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0319391 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.021188 | Absent |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.059719600000000005 | Absent |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.0192775 | Absent |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0482843 | Absent |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.00641441 | Absent |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.034833800000000005 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.00938264 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.0246329 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.006326300000000001 | Absent |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 0.0553877 | Absent |
| NPC;F129;A;GEO |
Significance: 0.0493776 | Absent |
| NPC;Ch13;A;GEO |
Significance: 0.00829637 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.0485491 | Absent |
| LNG;A;CL |
Significance: 0.014724700000000002 | Absent |
| Heart;D;En |
Significance: 0.016446900000000004 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.0350535 | Absent |
| BRN;PTMN;A;CL |
Significance: 0.0121798 | Absent |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.0463288 | Absent |
| LNG;8wks;D;GEO |
Significance: 0.0521226 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.0588839 | Absent |
| KDNY;P0;D;En |
Significance: 0.042651999999999995 | Absent |
| LNG;8wks;D;En |
Significance: 0.034971300000000004 | Absent |
| Heart;8wks;D;En |
Significance: 0.016851400000000002 | Absent |
| Heart;P0;D;En |
Significance: 0.030083299999999997 | Absent |
| wES;RTN;E11.5;D;En |
Significance: 0.0329357 | Absent |
| CRBLM;D;En |
Significance: 0.0533246 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.036423699999999996 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.0556206 | Absent |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.00907555 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.054964599999999995 | Absent |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.0404717 | Absent |
| WHL_BRN;8wks;D;En |
Significance: 0.0163973 | Absent |
Unique Cell types only Homer finds Snai1: #19
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En | Absent |
Significance: 0.032787800000000006 |
| ES;F1216;A;GEO | Absent |
Significance: 0.043979199999999996 |
| LNG;P0;D;En | Absent |
Significance: 0.050705400000000005 |
| wES;LNG;E14.5;D;En | Absent |
Significance: 0.0493234 |
| ES;ZhBTc4;2;D;GEO | Absent |
Significance: 0.0331605 |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL | Absent |
Significance: 0.0297182 |
| B_SPLND;A;GEO | Absent |
Significance: 0.030499900000000003 |
| aB_LPS;A;CL | Absent |
Significance: 0.054056799999999995 |
| DP3a;A;CL | Absent |
Significance: 0.0472962 |
| aB_LSP+a_d_dextran;A;CL | Absent |
Significance: 0.040854699999999994 |
| aB_CD40+IL4;A;CL | Absent |
Significance: 0.0359168 |
| gamma_delta_TCR_M-S-;A;CL | Absent |
Significance: 0.00892111 |
| immatureB_SPLND;CD43-_CD11b-;D;GEO | Absent |
Significance: 0.0503809 |
| Corticotrope;A;GEO | Absent |
Significance: 0.0480531 |
| NTRPHL;A;CL | Absent |
Significance: 0.0394164 |
| Muscle;A;CL | Absent |
Significance: 0.058948900000000005 |
| T_MLD;TCR_SV40-I_CD8+;D7;A;GEO | Absent |
Significance: 0.0486797 |
| Mammary_BptfKO;A;GEO | Absent |
Significance: 0.0142546 |
| wES;WHL_BRN;E14.5;D;En | Absent |
Significance: 0.041365 |