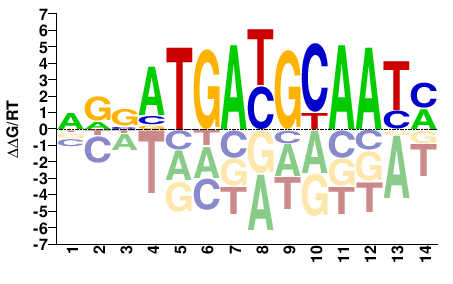
# Parameters
data_name = "Atf4"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 10:58:11,633 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Atf4: #39
| Cell type | Modisco | Homer |
|---|---|---|
| SKLTL_Muscle;8wks;D;En |
Significance: 0.05069 |
Significance: 0.055129899999999996 |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.0033106000000000003 |
Significance: 0.000403475 |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.047364699999999996 |
Significance: 0.0572279 |
| AML;D;En |
Significance: 0.0169911 |
Significance: 0.0196232 |
| DC_LPS;6h;A;GEO |
Significance: 0.00480388 |
Significance: 0.043023 |
| BSPHL;A;CL |
Significance: 0.00478372 |
Significance: 0.029245599999999997 |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 0.00891677 |
Significance: 0.0217183 |
| DC_LPS;2h;A;GEO |
Significance: 0.0188762 |
Significance: 0.057389499999999996 |
| NTRPHL;A;CL |
Significance: 0.0525279 |
Significance: 0.059038400000000005 |
| DC_pBMD;A;GEO |
Significance: 0.00662452 |
Significance: 0.00055455 |
| NIH3T3;D;En |
Significance: 0.00317009 |
Significance: 0.0530938 |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.0529155 |
Significance: 0.05405359999999999 |
| 3T3-L1;D;En |
Significance: 0.054037699999999994 |
Significance: 0.00298238 |
| MCRPHG;A;CL |
Significance: 0.0230418 |
Significance: 0.027051599999999995 |
| iPV_NRN;A;GEO |
Significance: 0.0414381 |
Significance: 0.058155899999999996 |
| Adrenal;A;CL |
Significance: 0.00974841 |
Significance: 0.044613599999999996 |
| placenta;A;CL |
Significance: 0.0204512 |
Significance: 0.033801599999999994 |
| pMeg;A;En |
Significance: 0.00553869 |
Significance: 0.0189835 |
| MegP;A;En |
Significance: 0.00386808 |
Significance: 0.029351099999999998 |
| DC_LPS;4h;A;En |
Significance: 0.046883600000000004 |
Significance: 0.0498324 |
| pGMP;A;En |
Significance: 0.00467485 |
Significance: 0.0216881 |
| DC;A;En |
Significance: 0.00364727 |
Significance: 0.050354699999999995 |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.00888805 |
Significance: 0.028654400000000003 |
| LVR;8wks;D;GEO |
Significance: 0.00879772 |
Significance: 0.0292207 |
| LNG;8wks;D;GEO |
Significance: 0.0111308 |
Significance: 0.0518668 |
| 416B_MP;CD34+;D;GEO |
Significance: 0.00346472 |
Significance: 0.0596342 |
| NIH3T3_FBROBLST;D;En |
Significance: 0.00526181 |
Significance: 0.058305899999999994 |
| FBRBLST;8wks;D;En |
Significance: 0.00557229 |
Significance: 0.059344900000000006 |
| 3T3-L1;FBRBLST;D;En |
Significance: 0.05777569999999999 |
Significance: 0.00036783199999999996 |
| LVR;8wks;D;En |
Significance: 0.00631426 |
Significance: 0.0422733 |
| NTRPHL;A;GEO |
Significance: 0.010059700000000001 |
Significance: 0.05499580000000001 |
| LVR;P0;D;En |
Significance: 0.0109089 |
Significance: 0.0468017 |
| LNG;8wks;D;En |
Significance: 0.00878896 |
Significance: 0.0272846 |
| LNG;P0;D;En |
Significance: 0.00507467 |
Significance: 0.0522255 |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 0.0108314 |
Significance: 0.0564624 |
| Adipose;D;En |
Significance: 0.058603800000000004 |
Significance: 0.0170097 |
| CRBLM;D;En |
Significance: 0.044493 |
Significance: 0.021243900000000003 |
| 416B_MP;CD34+;D;En |
Significance: 0.00748973 |
Significance: 0.0216956 |
| Muller;P12;D;En |
Significance: 0.00487533 |
Significance: 0.0437883 |
Unique Cell types only TF-MoDISco finds Atf4: #52
| Cell type | Modisco | Homer |
|---|---|---|
| MPP;A;CL |
Significance: 0.00621977 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.0531019 | Absent |
| LRG_INTSTN;8wks;D;En |
Significance: 0.0333294 | Absent |
| STMCH;P0;D;En |
Significance: 0.0514173 | Absent |
| 3134_mammaryADNCRCNM;D;En |
Significance: 0.0580356 | Absent |
| MCRPHG_BMD.LPD_Stmltd;A;GEO |
Significance: 0.00613499 | Absent |
| CH12.LX;D;En |
Significance: 0.029481599999999997 | Absent |
| AML;AE9a;A;GEO |
Significance: 0.05445030000000001 | Absent |
| MEL;D;En |
Significance: 0.0268976 | Absent |
| MEL_Znc_di-cl;24h;D;En |
Significance: 0.0174051 | Absent |
| ESNPHL_SPLND_BlbCA;CL |
Significance: 0.00718684 | Absent |
| MN1;D;En |
Significance: 0.05255309999999999 | Absent |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.05994730000000001 | Absent |
| SKN_EPTHL;A;GEO |
Significance: 0.028668700000000002 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.024882400000000002 | Absent |
| NPC;F129;A;GEO |
Significance: 0.0546184 | Absent |
| DC_BMD;A;CL |
Significance: 0.04783540000000001 | Absent |
| NPC;Ch13;A;GEO |
Significance: 0.056170000000000005 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.0433556 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.056134 | Absent |
| ES;MEF;A;CL |
Significance: 0.00028571900000000003 | Absent |
| ThMS_MEC;Aire++;A;GEO |
Significance: 0.016412700000000002 | Absent |
| Corticotrope;A;GEO |
Significance: 0.027034199999999998 | Absent |
| LNG;A;CL |
Significance: 0.0135915 | Absent |
| Muscle;A;CL |
Significance: 0.05410269999999999 | Absent |
| Heart;D;En |
Significance: 0.028540899999999998 | Absent |
| Melanotrope;A;GEO |
Significance: 0.0426441 | Absent |
| Mammary;A;CL |
Significance: 0.0530302 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.0543947 | Absent |
| Mammary_BptfKO;A;GEO |
Significance: 0.00359984 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.05544059999999999 | Absent |
| pCMP;A;CL |
Significance: 0.059949199999999994 | Absent |
| BRN;FRNTL_CRTX;A;CL |
Significance: 0.037266900000000006 | Absent |
| BRN;PTMN;A;CL |
Significance: 0.059602999999999996 | Absent |
| BRN;CRBLM;A;CL |
Significance: 0.0479684 | Absent |
| LNG_TRM_HSC;A;CL |
Significance: 0.0504218 | Absent |
| 3134_mammaryADNCRCNM;D;GE |
Significance: 0.0154423 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.0574485 | Absent |
| wCH12_drug;A;CL |
Significance: 4.82961e-05 | Absent |
| KDNY;P0;D;En |
Significance: 0.0323589 | Absent |
| KDNY;8wks;D;En |
Significance: 0.0542518 | Absent |
| wES;LVR;E11.5;D.;En |
Significance: 0.014094399999999998 | Absent |
| wES;LNG;E14.5;D;En |
Significance: 0.0230215 | Absent |
| ES;v6.5;A;GEO |
Significance: 0.05395369999999999 | Absent |
| Mammary_GLND;A;GEO |
Significance: 0.00280253 | Absent |
| Heart;8wks;D;En |
Significance: 0.023212599999999996 | Absent |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.05846669999999999 | Absent |
| Heart;P0;D;En |
Significance: 0.05949490000000001 | Absent |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.0477347 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.03773630000000001 | Absent |
| Md_BRN;D;En |
Significance: 0.054172500000000005 | Absent |
| WHL_BRN;8wks;D;En |
Significance: 0.0541928 | Absent |
Unique Cell types only Homer finds Atf4: #4
| Cell type | Modisco | Homer |
|---|---|---|
| restingB;A;CL | Absent |
Significance: 0.0587057 |
| ILC1_AbxGRMFree;A;GEO | Absent |
Significance: 0.05542419999999999 |
| ePyramidal_NRN;A;GEO | Absent |
Significance: 0.0458534 |
| aB_LPS+IL21;A;CL | Absent |
Significance: 0.0530492 |