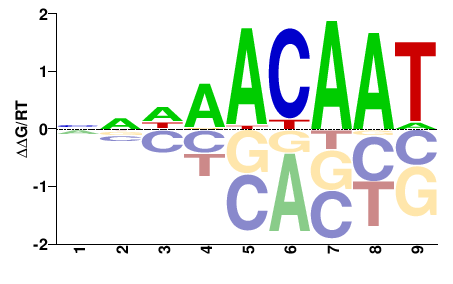
# Parameters
data_name = "Sox6"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:36:22,227 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Sox6: #21
| Cell type | Modisco | Homer |
|---|---|---|
| ENDRM_ESD;A;GEO |
Significance: 0.00396967 |
Significance: 0.00487707 |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00179107 |
Significance: 0.0011087999999999998 |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.00128714 |
Significance: 0.00404062 |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.00155 |
Significance: 0.00010964899999999999 |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0020921 |
Significance: 0.0159546 |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.0011604 |
Significance: 0.00252877 |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.00156284 |
Significance: 0.0289586 |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00122592 |
Significance: 0.000282777 |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.0007186 |
Significance: 0.0279811 |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.00169645 |
Significance: 0.000187963 |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.00257506 |
Significance: 0.0482267 |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.00129506 |
Significance: 0.00311784 |
| NPC;F129;A;GEO |
Significance: 0.00467444 |
Significance: 0.00524018 |
| NPC;Ch13;A;GEO |
Significance: 0.00070628 |
Significance: 0.014266299999999997 |
| ES;E14;A;CL |
Significance: 0.030060700000000003 |
Significance: 0.011410700000000001 |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.00751589 |
Significance: 0.052451099999999994 |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.0569535 |
Significance: 0.0161626 |
| ES;v6.5;A;GEO |
Significance: 0.008460700000000002 |
Significance: 0.0352176 |
| wES;RTN;E11.5;D;En |
Significance: 0.00161317 |
Significance: 0.017250099999999997 |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.00124326 |
Significance: 0.00122965 |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.00699687 |
Significance: 0.0184209 |
Unique Cell types only TF-MoDISco finds Sox6: #51
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En |
Significance: 0.00223121 | Absent |
| wES;limb;E11.5;D;En |
Significance: 0.0040207 | Absent |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.0243274 | Absent |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.00302883 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.00872091 | Absent |
| wES;FaceP;E11.5;D;En |
Significance: 0.00552292 | Absent |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.00172576 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.00384316 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.00394355 | Absent |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 0.0375099 | Absent |
| NPC;G4;A;GEO |
Significance: 0.00756561 | Absent |
| NPC;mE6;A;GEO |
Significance: 0.00129544 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.046486 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.00248838 | Absent |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.00458131 | Absent |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.0278991 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.014894399999999999 | Absent |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 0.021613900000000002 | Absent |
| Mammary;A;CL |
Significance: 0.0012195 | Absent |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.0437286 | Absent |
| Mammary_BptfKO;A;GEO |
Significance: 0.00134012 | Absent |
| SPNL_CHRD;A;CL |
Significance: 0.00290416 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.00732306 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.00371163 | Absent |
| BRN;PTMN;A;CL |
Significance: 0.0152393 | Absent |
| BRN;MDL_OBLNGT;A;CL |
Significance: 0.00743085 | Absent |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.00418211 | Absent |
| ES;D0;D;GEO |
Significance: 0.0119045 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.01757 | Absent |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.00950671 | Absent |
| ES;E14;D;En |
Significance: 0.0394088 | Absent |
| FBRBLST;8wks;D;En |
Significance: 0.00204365 | Absent |
| ES;J1;A;CL |
Significance: 0.019808700000000002 | Absent |
| ES;E14;A;GEO |
Significance: 0.0200705 | Absent |
| ES;F1216;A;GEO |
Significance: 0.0232785 | Absent |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.00565748 | Absent |
| Mammary_GLND;A;GEO |
Significance: 0.00355615 | Absent |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.026923900000000004 | Absent |
| Adipose;D;En |
Significance: 0.000640021 | Absent |
| RTN;D1;D;En |
Significance: 0.000128228 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 0.00223893 | Absent |
| Muller;P12;D;En |
Significance: 0.00255611 | Absent |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.00661102 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.00396898 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.00971682 | Absent |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.00338051 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.004553499999999999 | Absent |
| Md_BRN;D;En |
Significance: 0.00394229 | Absent |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.023208000000000003 | Absent |
| WHL_BRN;8wks;D;En |
Significance: 0.00390414 | Absent |
| wES;Telenchephalon;D;En |
Significance: 0.00309365 | Absent |