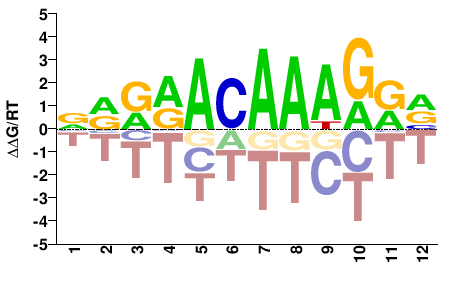
# Parameters
data_name = "Sox4"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:36:20,552 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Sox4: #21
| Cell type | Modisco | Homer |
|---|---|---|
| ENDRM_ESD;A;GEO |
Significance: 0.00674688 |
Significance: 0.029236500000000002 |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.006281 |
Significance: 0.00187683 |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.00888291 |
Significance: 0.0201657 |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.00728594 |
Significance: 0.00051925 |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0091527 |
Significance: 0.00670202 |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.00452083 |
Significance: 0.00293655 |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.0100903 |
Significance: 0.0289586 |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00732673 |
Significance: 0.00114587 |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.00560447 |
Significance: 0.0253197 |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.00938326 |
Significance: 0.00062266 |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.0138143 |
Significance: 0.051841 |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.0102737 |
Significance: 0.00369568 |
| NPC;F129;A;GEO |
Significance: 0.00747998 |
Significance: 0.027494099999999997 |
| NPC;Ch13;A;GEO |
Significance: 0.00318825 |
Significance: 0.0139929 |
| NPC;mE6;A;GEO |
Significance: 0.00552389 |
Significance: 0.0189149 |
| ES;E14;A;CL |
Significance: 0.0080676 |
Significance: 0.00206934 |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.0107973 |
Significance: 0.0037567 |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.00547122 |
Significance: 0.00990351 |
| wES;RTN;E11.5;D;En |
Significance: 0.00784392 |
Significance: 0.00501142 |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.00859767 |
Significance: 0.00211327 |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.0123243 |
Significance: 0.0286289 |
Unique Cell types only TF-MoDISco finds Sox4: #53
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En |
Significance: 0.00282207 | Absent |
| wES;limb;E11.5;D;En |
Significance: 0.0227202 | Absent |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.0510168 | Absent |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.0253696 | Absent |
| wES;FaceP;E14.5;D;En |
Significance: 0.0217241 | Absent |
| wES;FaceP;E11.5;D;En |
Significance: 0.024011599999999998 | Absent |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.0111242 | Absent |
| smallDPprecrsor;CD4+CD8+CD69-;A;CL |
Significance: 0.0487423 | Absent |
| CD4+CD8intTCR+CD69+CD24+;B2defM;A;CL |
Significance: 0.0549653 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.0286314 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.0500223 | Absent |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 0.00531263 | Absent |
| NPC;G4;A;GEO |
Significance: 0.0159309 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.00316299 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.00329485 | Absent |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.0178269 | Absent |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.0223138 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.0165061 | Absent |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 0.00742897 | Absent |
| Mammary;A;CL |
Significance: 0.00574347 | Absent |
| ES;JM8.N4;Mitosis;A;GEO |
Significance: 0.011164 | Absent |
| Mammary_BptfKO;A;GEO |
Significance: 0.00452942 | Absent |
| SPNL_CHRD;A;CL |
Significance: 0.0183864 | Absent |
| NRV;Eye-OPTC;A;CL |
Significance: 0.019172099999999997 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.0128291 | Absent |
| BRN;PTMN;A;CL |
Significance: 0.032793699999999995 | Absent |
| BRN;MDL_OBLNGT;A;CL |
Significance: 0.052629300000000004 | Absent |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.0168666 | Absent |
| ES;D0;D;GEO |
Significance: 0.0237088 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.0507983 | Absent |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.0210546 | Absent |
| ES;E14;D;En |
Significance: 0.039042 | Absent |
| FBRBLST;8wks;D;En |
Significance: 0.0074639 | Absent |
| ES;J1;A;CL |
Significance: 0.00316364 | Absent |
| ES;E14;A;GEO |
Significance: 0.038549099999999996 | Absent |
| ES;F1216;A;GEO |
Significance: 0.00203124 | Absent |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.015140200000000001 | Absent |
| ES;v6.5;A;GEO |
Significance: 0.0344262 | Absent |
| Mammary_GLND;A;GEO |
Significance: 0.00681352 | Absent |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.0409972 | Absent |
| Adipose;D;En |
Significance: 0.00131923 | Absent |
| RTN;D1;D;En |
Significance: 0.0034955 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 0.0161301 | Absent |
| Muller;P12;D;En |
Significance: 0.00667259 | Absent |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.0184473 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.0156827 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.0128161 | Absent |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.00994247 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.0165352 | Absent |
| Md_BRN;D;En |
Significance: 0.016016100000000002 | Absent |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.00438866 | Absent |
| WHL_BRN;8wks;D;En |
Significance: 0.0135286 | Absent |
| wES;Telenchephalon;D;En |
Significance: 0.0271597 | Absent |