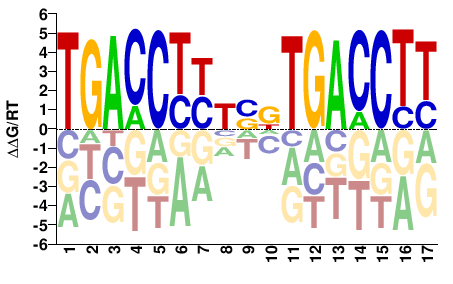
# Parameters
data_name = "Rarg"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:30:44,588 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Rarg: #20
| Cell type | Modisco | Homer |
|---|---|---|
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.00724174 |
Significance: 0.0379323 |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.05765750000000001 |
Significance: 0.0580435 |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.00791345 |
Significance: 0.0582553 |
| KDNY;8wks;D;En |
Significance: 0.0403837 |
Significance: 0.024848099999999998 |
| ES;E14;D;En |
Significance: 0.000207719 |
Significance: 0.0585054 |
| LVR;8wks;D;En |
Significance: 0.0212981 |
Significance: 0.0112038 |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.000188808 |
Significance: 0.0400087 |
| LRG_INTSTN;8wks;D;En |
Significance: 0.0363508 |
Significance: 0.027571 |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.000348895 |
Significance: 0.00300698 |
| ES;D0;D;GEO |
Significance: 2.6613099999999997e-06 |
Significance: 0.043118300000000005 |
| LVR;8wks;D;GEO |
Significance: 0.024915299999999998 |
Significance: 0.022453400000000002 |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 0.028305900000000002 |
Significance: 0.0588461 |
| ES;ZhBTc4;untreated;A;GEO |
Significance: 0.00153034 |
Significance: 0.00132601 |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.000314837 |
Significance: 0.00281218 |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.0378403 |
Significance: 0.00111077 |
| ES;C57BL_6J;R1;D;En |
Significance: 9.9462e-05 |
Significance: 0.00132819 |
| Adrenal;A;CL |
Significance: 0.00168537 |
Significance: 8.37478e-06 |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 4.1042e-05 |
Significance: 0.00209451 |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0542436 |
Significance: 0.0470511 |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.0129572 |
Significance: 0.043267599999999996 |
Unique Cell types only TF-MoDISco finds Rarg: #65
| Cell type | Modisco | Homer |
|---|---|---|
| T_MLD;TCR_SV40-I_CD8+;D28;A;GEO |
Significance: 0.0548582 | Absent |
| wES;limb;E14.5;D;En |
Significance: 0.058410800000000006 | Absent |
| memT;CD4+;A;CL |
Significance: 0.0398241 | Absent |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.0261276 | Absent |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.045490800000000005 | Absent |
| proB;A;CL |
Significance: 0.0473947 | Absent |
| memT_tmrD;TCR_SV40-I_CD8+;D7;A;GEO |
Significance: 0.0566149 | Absent |
| memT_tmrD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.05942240000000001 | Absent |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.058540999999999996 | Absent |
| STMCH;P0;D;En |
Significance: 0.023400099999999997 | Absent |
| prePLSM_B;A;GEO |
Significance: 0.0563621 | Absent |
| plasmaB;A;CL |
Significance: 0.0527028 | Absent |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.0157143 | Absent |
| BSPHL;A;CL |
Significance: 0.057650599999999996 | Absent |
| GCN_PHT_RCPTR;A;GEO |
Significance: 0.031931299999999996 | Absent |
| aNonTfh;A;CL |
Significance: 0.0342621 | Absent |
| aNonTfh_mut;A;CL |
Significance: 0.038069 | Absent |
| CN_PHT_RCPTR;A;GEO |
Significance: 0.054673400000000004 | Absent |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.0550606 | Absent |
| naiveT;CD4+_CD44-_CD62L-_CD25-;A;CL |
Significance: 0.029937799999999997 | Absent |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0194509 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.053231600000000004 | Absent |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.031020999999999996 | Absent |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.0027523000000000005 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.053682400000000005 | Absent |
| CD4+CD8intTCR+CD69+CD24+;MHC2defM;A;CL |
Significance: 0.053829999999999996 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.014282399999999999 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.028584699999999998 | Absent |
| CD4+CD8-TCR+CD25+Foxp3-;A;CL |
Significance: 0.0550452 | Absent |
| CD4+CD8-TCR+CD25-Foxp3+;A;CL |
Significance: 0.0587678 | Absent |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL |
Significance: 0.048833999999999995 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.0474733 | Absent |
| ES;E14;A;CL |
Significance: 0.0127479 | Absent |
| SML_INTSTN;A;CL |
Significance: 0.0554733 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.0136211 | Absent |
| ThMS_MEC;Aire++;A;GEO |
Significance: 0.011742299999999999 | Absent |
| Muscle;A;CL |
Significance: 0.0420103 | Absent |
| Melanotrope;A;GEO |
Significance: 0.05986469999999999 | Absent |
| ES;JM8.N4;Asynch;A;GEO |
Significance: 0.057446000000000004 | Absent |
| placenta;A;CL |
Significance: 0.054093499999999996 | Absent |
| pMeg;A;En |
Significance: 0.0463009 | Absent |
| eye-NRL_RTN;A;CL |
Significance: 0.0281714 | Absent |
| MegP;A;En |
Significance: 0.051361300000000006 | Absent |
| BRN;CRBLM;A;CL |
Significance: 0.035442400000000006 | Absent |
| gamma_delta_TCR;A;CL |
Significance: 0.040525099999999994 | Absent |
| T_SPLND_blstCLTRD;CD4+;D;GEO |
Significance: 0.058122400000000005 | Absent |
| apCD4Eff;D;GEO |
Significance: 0.0551668 | Absent |
| 416B_MP;CD34+;D;GEO |
Significance: 5.96395e-05 | Absent |
| CH12_QNTPLE;A;CL |
Significance: 0.0520058 | Absent |
| KDNY;P0;D;En |
Significance: 0.0468558 | Absent |
| mem_precrsrT_SPLND;CD8+;A;GEO |
Significance: 0.0533773 | Absent |
| wES;LVR;E11.5;D.;En |
Significance: 0.051754999999999995 | Absent |
| ES;J1;A;CL |
Significance: 0.05899069999999999 | Absent |
| LVR;P0;D;En |
Significance: 0.0418066 | Absent |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.000251542 | Absent |
| Heart;8wks;D;En |
Significance: 0.050388199999999994 | Absent |
| Heart;P0;D;En |
Significance: 0.059919900000000005 | Absent |
| RTN;A;GEO |
Significance: 0.035430800000000005 | Absent |
| ILC2+IL33+IL25;A;GEO |
Significance: 0.057447000000000005 | Absent |
| 416B_MP;CD34+;D;En |
Significance: 0.00022735 | Absent |
| T_LCMV;CD8+;D33;A;GEO |
Significance: 0.04391180000000001 | Absent |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.0574821 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 0.025366299999999998 | Absent |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.0566153 | Absent |
| cntrl_memT;TCR_SV40-I_CD8+;A;GEO |
Significance: 0.0371175 | Absent |
Unique Cell types only Homer finds Rarg: #2
| Cell type | Modisco | Homer |
|---|---|---|
| pCMP;A;CL | Absent |
Significance: 0.059524400000000005 |
| DC_BMD;A;CL | Absent |
Significance: 0.029620999999999998 |