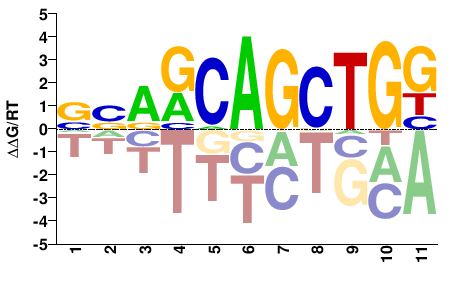
# Parameters
data_name = "Bhlha15"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 10:59:10,962 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Bhlha15: #39
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En |
Significance: 0.0169932 |
Significance: 0.013133500000000001 |
| wES;limb;E11.5;D;En |
Significance: 0.00770948 |
Significance: 0.052213999999999997 |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.018165200000000003 |
Significance: 0.0263493 |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.048054599999999996 |
Significance: 0.048942900000000004 |
| wES;FaceP;E14.5;D;En |
Significance: 0.0410005 |
Significance: 0.0316694 |
| wES;FaceP;E11.5;D;En |
Significance: 0.0165506 |
Significance: 0.0532775 |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.015087999999999999 |
Significance: 0.0241832 |
| SKLTL_Muscle;8wks;D;En |
Significance: 0.010476000000000001 |
Significance: 0.028565499999999997 |
| plasmaB;A;CL |
Significance: 0.00133412 |
Significance: 0.022834200000000002 |
| preB_BMD;A;GEO |
Significance: 0.00020360900000000002 |
Significance: 0.000335661 |
| B;B220_CD43;A;GEO |
Significance: 0.000599735 |
Significance: 0.014593700000000001 |
| 3T3-L1;D;En |
Significance: 0.0349612 |
Significance: 0.056063699999999994 |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 0.017681 |
Significance: 0.0405639 |
| NPC;F129;A;GEO |
Significance: 0.00582295 |
Significance: 0.0159364 |
| NPC;G4;A;GEO |
Significance: 0.000617165 |
Significance: 0.00815796 |
| NPC;Ch13;A;GEO |
Significance: 0.00855318 |
Significance: 0.00689822 |
| NPC;mE6;A;GEO |
Significance: 0.00142326 |
Significance: 0.0115942 |
| ePyramidal_NRN;A;GEO |
Significance: 0.023866099999999998 |
Significance: 0.00247762 |
| eCortical_NRN;A;GEO |
Significance: 0.0286909 |
Significance: 0.00077762 |
| G_NRN_P;A;GEO |
Significance: 0.00697041 |
Significance: 0.0253346 |
| Heart;D;En |
Significance: 0.0186097 |
Significance: 0.056486800000000004 |
| BRN;FRNTL_CRTX;A;CL |
Significance: 0.0152666 |
Significance: 0.00047420599999999997 |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.012074 |
Significance: 0.0055931999999999996 |
| BRN;CRBLM;A;CL |
Significance: 0.018481599999999997 |
Significance: 0.0182432 |
| WHL_BRN;8wks;D;GEO |
Significance: 0.0294981 |
Significance: 0.0230692 |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.00703216 |
Significance: 0.00110117 |
| KDNY;P0;D;En |
Significance: 0.054024800000000005 |
Significance: 0.0575499 |
| LNG;P0;D;En |
Significance: 0.0412994 |
Significance: 0.0155107 |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.00532126 |
Significance: 0.00263517 |
| RTN;D1;D;En |
Significance: 0.026290499999999998 |
Significance: 0.011891200000000001 |
| wES;RTN;E11.5;D;En |
Significance: 0.014379399999999999 |
Significance: 0.058075699999999994 |
| wES;MSDRM;E11.5;D;En |
Significance: 0.0110114 |
Significance: 0.00308512 |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.00811636 |
Significance: 0.03175 |
| Fr_BRN;P0;D;En |
Significance: 0.0311348 |
Significance: 0.00267641 |
| Hnd_BRN;P0;D;En |
Significance: 0.011445100000000001 |
Significance: 0.028760400000000005 |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.0347133 |
Significance: 0.0324823 |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.00394079 |
Significance: 0.0179583 |
| WHL_BRN;8wks;D;En |
Significance: 0.0304343 |
Significance: 0.0339913 |
| wES;Telenchephalon;D;En |
Significance: 0.0263507 |
Significance: 0.0194387 |
Unique Cell types only TF-MoDISco finds Bhlha15: #43
| Cell type | Modisco | Homer |
|---|---|---|
| proB;A;CL |
Significance: 0.000668353 | Absent |
| preB;A;CL |
Significance: 0.000672082 | Absent |
| B1_B;A;CL |
Significance: 0.04648819999999999 | Absent |
| prePLSM_B;A;GEO |
Significance: 0.00784307 | Absent |
| NIH3T3;D;En |
Significance: 0.0159248 | Absent |
| GCN_PHT_RCPTR;A;GEO |
Significance: 0.0307514 | Absent |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.0238531 | Absent |
| CN_PHT_RCPTR;A;GEO |
Significance: 0.0518774 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.019953099999999998 | Absent |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.045538300000000004 | Absent |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.025775799999999998 | Absent |
| largeDPprecrsr;CD4+CD8+CD69-;A;CL |
Significance: 0.0212349 | Absent |
| smallDPprecrsor;CD4+CD8+CD69-;A;CL |
Significance: 0.030724 | Absent |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.025777800000000003 | Absent |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00998294 | Absent |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.00600887 | Absent |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.014566899999999999 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.014057399999999998 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.0142395 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.00982 | Absent |
| DP;A;CL |
Significance: 0.0465283 | Absent |
| iPV_NRN;A;GEO |
Significance: 0.013796899999999999 | Absent |
| DP3b;A;CL |
Significance: 0.0337202 | Absent |
| ISP;A;CL |
Significance: 0.00202878 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.0337914 | Absent |
| Corticotrope;A;GEO |
Significance: 0.00304366 | Absent |
| LNG;A;CL |
Significance: 0.000574768 | Absent |
| Muscle;A;CL |
Significance: 0.010155299999999999 | Absent |
| Melanotrope;A;GEO |
Significance: 0.00506844 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.000509239 | Absent |
| NIH3T3_FBROBLST;D;GEO |
Significance: 0.0332898 | Absent |
| NIH3T3_FBROBLST;D;En |
Significance: 0.018043599999999996 | Absent |
| FBRBLST;8wks;D;En |
Significance: 0.05449059999999999 | Absent |
| wCH12_drug;A;CL |
Significance: 0.00544895 | Absent |
| 3T3-L1;FBRBLST;D;En |
Significance: 0.0424763 | Absent |
| LNG;8wks;D;En |
Significance: 0.00602435 | Absent |
| Heart;8wks;D;En |
Significance: 0.0583815 | Absent |
| RTN;A;GEO |
Significance: 0.048498599999999996 | Absent |
| CRBLM;D;En |
Significance: 0.017283700000000003 | Absent |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.00956437 | Absent |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.00428863 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.00474469 | Absent |
| Md_BRN;D;En |
Significance: 0.016348400000000003 | Absent |
Unique Cell types only Homer finds Bhlha15: #2
| Cell type | Modisco | Homer |
|---|---|---|
| wES;LNG;E14.5;D;En | Absent |
Significance: 0.0118503 |
| DP3a;A;CL | Absent |
Significance: 0.00959585 |