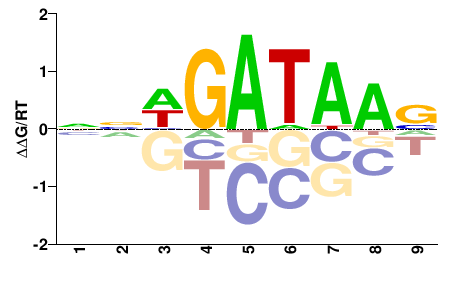
# Parameters
data_name = "Gata5"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:09:57,571 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Gata5: #15
| Cell type | Modisco | Homer |
|---|---|---|
| MegP;A;En |
Significance: 3.62085e-06 |
Significance: 0.0394318 |
| pCMP;A;CL |
Significance: 0.000225305 |
Significance: 0.00588901 |
| LVR;P0;D;En |
Significance: 1.34402e-05 |
Significance: 0.00254251 |
| wES;LVR;E11.5;D.;En |
Significance: 0.000210425 |
Significance: 0.00820501 |
| Heart;P0;D;En |
Significance: 0.0018482 |
Significance: 0.0043076 |
| STMCH;P0;D;En |
Significance: 0.0226202 |
Significance: 0.047006 |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.000445988 |
Significance: 0.015191899999999998 |
| Erythroid;Ter119+;A;GEO |
Significance: 9.56579e-06 |
Significance: 0.000688864 |
| AML;AE9a;A;GEO |
Significance: 0.00275473 |
Significance: 0.00040374 |
| Mast;A;CL |
Significance: 0.0352471 |
Significance: 0.0377097 |
| BSPHL;A;CL |
Significance: 0.0344851 |
Significance: 0.0297543 |
| 416B_MP;CD34+;D;GEO |
Significance: 0.0033710000000000003 |
Significance: 0.015570600000000002 |
| MEF.GATA-1-ER;D;En |
Significance: 0.000294541 |
Significance: 0.0313682 |
| MEL;D;En |
Significance: 0.000135483 |
Significance: 6.20455e-07 |
| MegEP;A;CL |
Significance: 0.00107627 |
Significance: 0.024049099999999997 |
Unique Cell types only TF-MoDISco finds Gata5: #32
| Cell type | Modisco | Homer |
|---|---|---|
| proB;A;CL |
Significance: 0.000189745 | Absent |
| B;B220_CD43;A;GEO |
Significance: 0.000482711 | Absent |
| ESNPHL_ex-vivo_BlbC;A;CL |
Significance: 0.00273108 | Absent |
| ESNPHL_SPLND_BlbCA;CL |
Significance: 0.00485036 | Absent |
| ENDRM_ESD;A;GEO |
Significance: 0.00623762 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.0549867 | Absent |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.00515322 | Absent |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.00157886 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.00573815 | Absent |
| SKN_EPTHL;A;GEO |
Significance: 0.0120624 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.0131626 | Absent |
| Erythroblast;A;CL |
Significance: 4.84705e-05 | Absent |
| SML_INTSTN;A;CL |
Significance: 0.036381300000000005 | Absent |
| LRG_INTSTN;A;CL |
Significance: 0.0233996 | Absent |
| Heart;D;En |
Significance: 0.00279696 | Absent |
| placenta;A;CL |
Significance: 0.00831743 | Absent |
| EPP;A;En |
Significance: 1.34541e-05 | Absent |
| pErythroblast;A;En |
Significance: 0.00010396799999999999 | Absent |
| LVR;8wks;D;GEO |
Significance: 0.00912097 | Absent |
| LNG_TRM_HSC;A;CL |
Significance: 8.987229999999999e-05 | Absent |
| SHRT_TRM_HSC;A;CL |
Significance: 0.000189291 | Absent |
| LNG;8wks;D;GEO |
Significance: 0.00943472 | Absent |
| T_SPLND_blstCLTRD;CD4+;D;GEO |
Significance: 0.053014 | Absent |
| NCR+ILC3+IL23;A;CL |
Significance: 0.015178899999999999 | Absent |
| ILC2;A;CL |
Significance: 0.0326358 | Absent |
| Th2_nippo;A;CL |
Significance: 0.0372356 | Absent |
| LNG;8wks;D;En |
Significance: 0.00918431 | Absent |
| Heart;8wks;D;En |
Significance: 0.00601254 | Absent |
| ILC2+IL33+IL25;A;GEO |
Significance: 0.0336362 | Absent |
| ILC2_AbxGRMFree;A;GEO |
Significance: 0.019997400000000002 | Absent |
| EP;c-kit+CD71+Ter119+;D;En |
Significance: 0.000195197 | Absent |
| 416B_MP;CD34+;D;En |
Significance: 0.000390596 | Absent |
Unique Cell types only Homer finds Gata5: #1
| Cell type | Modisco | Homer |
|---|---|---|
| MEL_Znc_di-cl;24h;D;En | Absent |
Significance: 0.019408900000000003 |