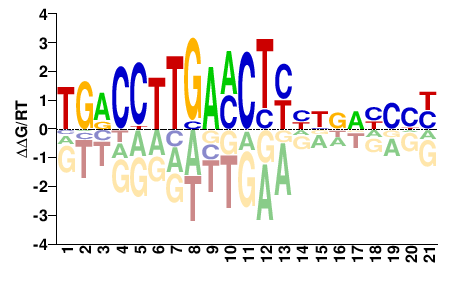
# Parameters
data_name = "Rxra"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:33:10,080 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Rxra: #25
| Cell type | Modisco | Homer |
|---|---|---|
| LRG_INTSTN;8wks;D;En |
Significance: 1.25942e-05 |
Significance: 0.000366395 |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0194509 |
Significance: 3.71626e-07 |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.053231600000000004 |
Significance: 0.0448941 |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.0542436 |
Significance: 0.03524 |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.0146909 |
Significance: 0.030509699999999997 |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.00660366 |
Significance: 0.0391305 |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0460619 |
Significance: 7.59551e-07 |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.00555084 |
Significance: 0.048524300000000006 |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.0572819 |
Significance: 0.029793200000000002 |
| SML_INTSTN;A;CL |
Significance: 1.10026e-05 |
Significance: 0.00626326 |
| LRG_INTSTN;A;CL |
Significance: 4.51962e-05 |
Significance: 0.00531345 |
| npES;ZHBTC4;doxyc;treat;A;GEO |
Significance: 0.000217455 |
Significance: 0.00506632 |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.0317265 |
Significance: 0.0510919 |
| Adrenal;A;CL |
Significance: 0.000343055 |
Significance: 2.00159e-05 |
| placenta;A;CL |
Significance: 0.054093499999999996 |
Significance: 1.1398299999999999e-08 |
| ES;ZhBTc4;1;D;GEO |
Significance: 0.00814486 |
Significance: 0.029466700000000002 |
| ES;ZhBTc4;2;D;GEO |
Significance: 0.000136738 |
Significance: 0.00420516 |
| ES;D0;D;GEO |
Significance: 0.014266399999999999 |
Significance: 0.00899205 |
| LVR;8wks;D;GEO |
Significance: 5.32854e-07 |
Significance: 3.4483499999999996e-05 |
| ES;129S1_SVImJ_CJ7;D;En |
Significance: 0.0479774 |
Significance: 0.002876 |
| ES;C57BL_6J;R1;D;En |
Significance: 0.015365100000000001 |
Significance: 0.000371962 |
| KDNY;8wks;D;En |
Significance: 0.052090599999999994 |
Significance: 9.599120000000001e-05 |
| LVR;8wks;D;En |
Significance: 8.444370000000001e-07 |
Significance: 2.46488e-05 |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 4.864560000000001e-05 |
Significance: 6.42241e-05 |
| wES;MSDRM;E11.5;D;En |
Significance: 0.015616399999999999 |
Significance: 4.21619e-07 |
Unique Cell types only TF-MoDISco finds Rxra: #31
| Cell type | Modisco | Homer |
|---|---|---|
| T_MLD;TCR_SV40-I_CD8+;D28;A;GEO |
Significance: 0.0433176 | Absent |
| wES;limb;E11.5;D;En |
Significance: 0.058220100000000004 | Absent |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.0198445 | Absent |
| memT_tmrD_lstr;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.0252319 | Absent |
| memT_tmrD;TCR_SV40-I_CD8+;D14;A;GEO |
Significance: 0.05942240000000001 | Absent |
| STMCH;P0;D;En |
Significance: 0.00548274 | Absent |
| 416B;HMTP_SPNSN;CD34+;D;En |
Significance: 0.0331756 | Absent |
| MEL;D;En |
Significance: 0.0546433 | Absent |
| Tfr_mut;CD4+CD44+CXCR5+PD1+CD25+;A;CL |
Significance: 0.0579774 | Absent |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.00776046 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.0505392 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.00296889 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.0307166 | Absent |
| CD4+CD8-TCR+CD25-Foxp3+;A;CL |
Significance: 0.043972699999999997 | Absent |
| CD4+CD8-TCR+CD25+Foxp3+;A;CL |
Significance: 0.0520505 | Absent |
| ThMS_MEC;Aire++;A;GEO |
Significance: 0.00237246 | Absent |
| Muscle;A;CL |
Significance: 0.0367901 | Absent |
| pCMP;A;CL |
Significance: 0.0345097 | Absent |
| 416B_MP;CD34+;D;GEO |
Significance: 0.00748905 | Absent |
| ES;E14;D;En |
Significance: 0.010334999999999999 | Absent |
| KDNY;P0;D;En |
Significance: 0.00020060599999999998 | Absent |
| wES;LVR;E11.5;D.;En |
Significance: 0.044206800000000004 | Absent |
| ES;J1;A;CL |
Significance: 0.0222415 | Absent |
| LVR;P0;D;En |
Significance: 0.054486400000000004 | Absent |
| ES;129SV_Jae_C57BL6J;A;GEO |
Significance: 0.0249739 | Absent |
| Heart;8wks;D;En |
Significance: 0.0284946 | Absent |
| Heart;P0;D;En |
Significance: 0.00671941 | Absent |
| naiveT;CD8+;A;GEO |
Significance: 0.0528096 | Absent |
| 416B_MP;CD34+;D;En |
Significance: 2.6458300000000003e-07 | Absent |
| T_LCMV;CD8+;D35;A;GEO |
Significance: 0.040009300000000005 | Absent |
| naiveT;trnsgncTCR_SV40-I_CD8;A;GEO |
Significance: 0.033262599999999996 | Absent |
Unique Cell types only Homer finds Rxra: #2
| Cell type | Modisco | Homer |
|---|---|---|
| DC_BMD;A;CL | Absent |
Significance: 0.026607799999999997 |
| ES;ZhBTc4;untreated;A;GEO | Absent |
Significance: 0.053002 |