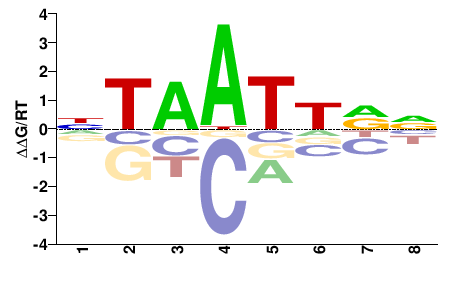
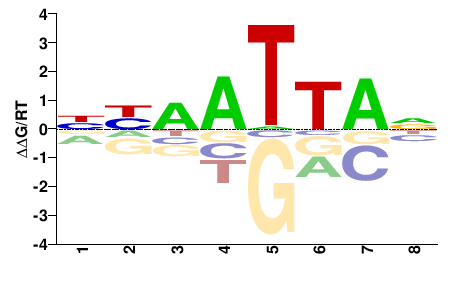
# Parameters
data_name = "Uncx"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:46:40,523 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Uncx: #24
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E11.5;D;En |
Significance: 0.00283111 |
Significance: 0.0529923 |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.00208611 |
Significance: 0.0349095 |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.00186405 |
Significance: 0.044849400000000005 |
| wES;FaceP;E14.5;D;En |
Significance: 0.0319054 |
Significance: 0.0578678 |
| wES;FaceP;E11.5;D;En |
Significance: 0.038419699999999994 |
Significance: 0.0356255 |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.0441381 |
Significance: 0.026036200000000002 |
| MN1;D;En |
Significance: 0.0544754 |
Significance: 0.0387827 |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.0317996 |
Significance: 0.024104499999999997 |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.0059272 |
Significance: 0.021843099999999997 |
| BCK_SKN;A;GEO |
Significance: 0.00257369 |
Significance: 0.021100999999999998 |
| NPC;F129;A;GEO |
Significance: 0.059852999999999996 |
Significance: 0.0409827 |
| iVIP_NRN;A;GEO |
Significance: 0.0496346 |
Significance: 0.037588199999999995 |
| G_NRN_P;A;GEO |
Significance: 0.00235131 |
Significance: 0.0218315 |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.0017565999999999999 |
Significance: 0.0579871 |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.00446988 |
Significance: 0.0211703 |
| RTN;D1;D;En |
Significance: 0.00342275 |
Significance: 0.024255099999999998 |
| wES;RTN;E11.5;D;En |
Significance: 0.00265193 |
Significance: 0.036596300000000005 |
| Muller;P12;D;En |
Significance: 0.00349225 |
Significance: 0.010803299999999998 |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.00277091 |
Significance: 0.036632300000000007 |
| Fr_BRN;P0;D;En |
Significance: 0.046874 |
Significance: 0.029008799999999998 |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.0265438 |
Significance: 0.033429900000000005 |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.00101197 |
Significance: 0.0226359 |
| Md_BRN;D;En |
Significance: 0.00918597 |
Significance: 0.00965481 |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.00212061 |
Significance: 0.057590499999999996 |
Unique Cell types only TF-MoDISco finds Uncx: #17
| Cell type | Modisco | Homer |
|---|---|---|
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.0502942 | Absent |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00837683 | Absent |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.00398197 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.00328194 | Absent |
| ES;E14;A;GEO |
Significance: 0.038042599999999996 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.00560467 | Absent |
| ES;v6.5;A;GEO |
Significance: 0.057272699999999996 | Absent |
| NPC;G4;A;GEO |
Significance: 0.054088300000000006 | Absent |
| NPC;mE6;A;GEO |
Significance: 0.036853199999999996 | Absent |
| iPV_NRN;A;GEO |
Significance: 0.044069699999999996 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 0.026727299999999996 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.00707871 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.056216699999999994 | Absent |
| ES;C57BL_6J;R1;D;En |
Significance: 0.0496519 | Absent |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00378015 | Absent |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.00995462 | Absent |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.0346034 | Absent |
Unique Cell types only Homer finds Uncx: #4
| Cell type | Modisco | Homer |
|---|---|---|
| wES.CRNL_NRL_CRST;E10.5;A;GEO | Absent |
Significance: 0.031042700000000003 |
| CN_PHT_RCPTR;A;GEO | Absent |
Significance: 0.022390900000000002 |
| BRN;OLFTRY_BLB;A;CL | Absent |
Significance: 0.0584003 |
| Melanotrope;A;GEO | Absent |
Significance: 0.0258511 |