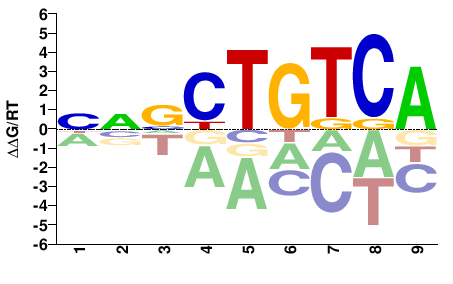
# Parameters
data_name = "Tgif1"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:45:23,240 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Tgif1: #19
| Cell type | Modisco | Homer |
|---|---|---|
| wES;FaceP;E14.5;D;En |
Significance: 0.0568121 |
Significance: 0.032621199999999996 |
| proB;A;CL |
Significance: 0.00637636 |
Significance: 0.0112068 |
| preB;A;CL |
Significance: 0.025961 |
Significance: 0.0102829 |
| BRN;PTMN;A;CL |
Significance: 0.00462071 |
Significance: 0.0494985 |
| Heart;8wks;D;En |
Significance: 0.0249329 |
Significance: 0.00573459 |
| SKLTL_Muscle;8wks;D;En |
Significance: 0.0193255 |
Significance: 0.00738291 |
| NPC;F129;A;GEO |
Significance: 0.019671 |
Significance: 0.014336700000000003 |
| NPC;G4;A;GEO |
Significance: 0.0468986 |
Significance: 0.00721965 |
| plasmaB;A;CL |
Significance: 0.00524925 |
Significance: 0.0113916 |
| preB_BMD;A;GEO |
Significance: 0.00728535 |
Significance: 0.0110289 |
| B;B220_CD43;A;GEO |
Significance: 0.0141344 |
Significance: 0.00894215 |
| NPC;Ch13;A;GEO |
Significance: 0.052459500000000006 |
Significance: 0.00567272 |
| NPC;mE6;A;GEO |
Significance: 0.027662799999999998 |
Significance: 0.0485226 |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.04846280000000001 |
Significance: 0.0167764 |
| RTN;D1;D;En |
Significance: 0.0313481 |
Significance: 0.0152315 |
| wES;MSDRM;E11.5;D;En |
Significance: 0.040922599999999996 |
Significance: 0.00907911 |
| Muscle;A;CL |
Significance: 0.00947066 |
Significance: 0.0428264 |
| Heart;D;En |
Significance: 0.00769393 |
Significance: 0.0144249 |
| wES;Md_BRN;E11.5;D;En |
Significance: 0.014763900000000002 |
Significance: 0.0267995 |
Unique Cell types only TF-MoDISco finds Tgif1: #31
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E11.5;D;En |
Significance: 0.0197491 | Absent |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.0327215 | Absent |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.044338199999999994 | Absent |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0201622 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.00486298 | Absent |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.00847325 | Absent |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.014842900000000001 | Absent |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.0038459 | Absent |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0452741 | Absent |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.014020099999999999 | Absent |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.0191048 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.00508457 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.0452962 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.00932805 | Absent |
| ISP;A;CL |
Significance: 0.0185857 | Absent |
| LNG;A;CL |
Significance: 0.0367615 | Absent |
| Melanotrope;A;GEO |
Significance: 0.0463666 | Absent |
| BRN;OLFTRY_BLB;A;CL |
Significance: 0.0349329 | Absent |
| LNG;8wks;D;GEO |
Significance: 0.0505402 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.018974 | Absent |
| wCH12_drug;A;CL |
Significance: 0.0337171 | Absent |
| KDNY;P0;D;En |
Significance: 0.0436659 | Absent |
| Heart;P0;D;En |
Significance: 0.0327039 | Absent |
| wES;RTN;E11.5;D;En |
Significance: 0.0510148 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.0162714 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.0504011 | Absent |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.0110867 | Absent |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.0424176 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.00846623 | Absent |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.0173813 | Absent |
| WHL_BRN;8wks;D;En |
Significance: 0.0240327 | Absent |
Unique Cell types only Homer finds Tgif1: #7
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En | Absent |
Significance: 0.0213429 |
| LNG;P0;D;En | Absent |
Significance: 0.0503891 |
| wES;LNG;E14.5;D;En | Absent |
Significance: 0.016567500000000002 |
| prePLSM_B;A;GEO | Absent |
Significance: 0.023848599999999998 |
| DP3a;A;CL | Absent |
Significance: 0.00889657 |
| iVIP_NRN;A;GEO | Absent |
Significance: 0.05508480000000001 |
| G_NRN_P;A;GEO | Absent |
Significance: 0.045372699999999995 |