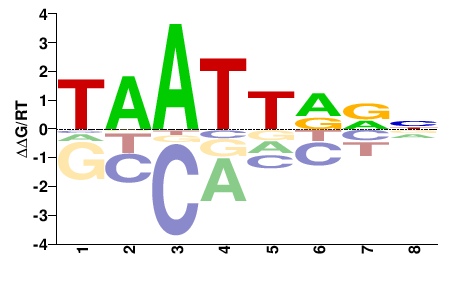
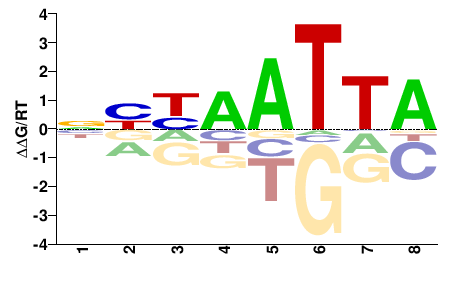
# Parameters
data_name = "Prrx1"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:30:09,276 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Prrx1: #22
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E11.5;D;En |
Significance: 0.00503938 |
Significance: 0.00824027 |
| wES;Fr_lmb;E11.5;D;En |
Significance: 0.00322653 |
Significance: 0.0413234 |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 0.00302253 |
Significance: 0.00218813 |
| wES;FaceP;E14.5;D;En |
Significance: 0.029881 |
Significance: 0.022596599999999998 |
| BL_CN_PHT_RCPTR;A;GEO |
Significance: 0.016986900000000003 |
Significance: 0.0093836 |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.0113635 |
Significance: 0.00986466 |
| BCK_SKN;A;GEO |
Significance: 0.00497263 |
Significance: 0.00314499 |
| NPC;F129;A;GEO |
Significance: 0.0494977 |
Significance: 0.0394772 |
| iVIP_NRN;A;GEO |
Significance: 0.043206699999999994 |
Significance: 0.010600299999999998 |
| G_NRN_P;A;GEO |
Significance: 0.00188899 |
Significance: 0.00719931 |
| Melanotrope;A;GEO |
Significance: 0.05850930000000001 |
Significance: 0.0117241 |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.0036217 |
Significance: 0.0019278 |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.00181216 |
Significance: 0.00669593 |
| RTN;D1;D;En |
Significance: 0.00321646 |
Significance: 0.00237484 |
| wES;RTN;E11.5;D;En |
Significance: 0.00232032 |
Significance: 0.00230796 |
| Muller;P12;D;En |
Significance: 0.0036969999999999998 |
Significance: 0.00310006 |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.0038116 |
Significance: 0.00161497 |
| Fr_BRN;P0;D;En |
Significance: 0.00476669 |
Significance: 0.00940302 |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.0476713 |
Significance: 0.00416828 |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.00288396 |
Significance: 0.0112762 |
| Md_BRN;D;En |
Significance: 0.0093618 |
Significance: 0.00433512 |
| wES;WHL_BRN;E14.5;D;En |
Significance: 0.00356192 |
Significance: 0.00218371 |
Unique Cell types only TF-MoDISco finds Prrx1: #18
| Cell type | Modisco | Homer |
|---|---|---|
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.00379006 | Absent |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00680081 | Absent |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.00820188 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.00567046 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.03337 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.00631577 | Absent |
| ES;E14;A;GEO |
Significance: 0.038042599999999996 | Absent |
| ES;v6.5;A;GEO |
Significance: 0.0399621 | Absent |
| NPC;G4;A;GEO |
Significance: 0.022274799999999997 | Absent |
| NPC;mE6;A;GEO |
Significance: 0.0317732 | Absent |
| iPV_NRN;A;GEO |
Significance: 0.0134408 | Absent |
| wES;MSDRM;E11.5;D;En |
Significance: 0.0271925 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.0492485 | Absent |
| Hnd_BRN;P0;D;En |
Significance: 0.00202336 | Absent |
| npES;ZHBTC4;Brg1fl_fl;untreat;A;GEO |
Significance: 0.0355852 | Absent |
| ES;C57BL_6J;R1;D;En |
Significance: 0.0341902 | Absent |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.00162683 | Absent |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.00132355 | Absent |
Unique Cell types only Homer finds Prrx1: #4
| Cell type | Modisco | Homer |
|---|---|---|
| wES.CRNL_NRL_CRST;E10.5;A;GEO | Absent |
Significance: 0.0369695 |
| CN_PHT_RCPTR;A;GEO | Absent |
Significance: 0.00868799 |
| BRN;OLFTRY_BLB;A;CL | Absent |
Significance: 0.0230635 |
| wES;FaceP;E11.5;D;En | Absent |
Significance: 0.0427584 |