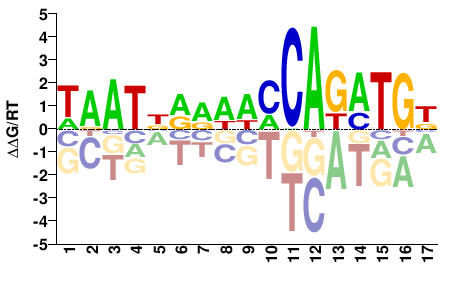
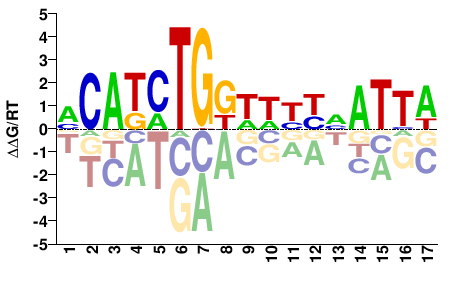
# Parameters
data_name = "Twist1"
task_idxs = "0-213,216-278"
homer_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_scans"
modisco_root = "/srv/scratch/msharmin/mouse_hem/with_tfd/full_mouse50/Naive_modisco2019"
overwrite = False
from matlas.reports import collect_motifDB_to_be_displayed, display_comparative_cell_types
load data from labcluster
Using TensorFlow backend. 2019-08-23 11:46:31,301 [WARNING] git-lfs not installed
homer_matches, modisco_matches = collect_motifDB_to_be_displayed(task_idxs, homer_root, modisco_root,
overwrite=False)
TF-MoDISco is using the TensorFlow backend.
display_comparative_cell_types(data_name, homer_matches, modisco_matches)
Shared Cell types both TF-MoDISco and Homer finds Twist1: #17
| Cell type | Modisco | Homer |
|---|---|---|
| wES;limb;E14.5;D;En |
Significance: 2.0155599999999998e-18 |
Significance: 0.00730577 |
| wES;limb;E11.5;D;En |
Significance: 8.3018e-05 |
Significance: 0.00016136 |
| wES;Fr_lmb;E11.5;D;En |
Significance: 1.23435e-07 |
Significance: 9.042229999999999e-05 |
| wES;Hnd_lmb;E11.5;D;En |
Significance: 5.88523e-18 |
Significance: 0.00016051799999999998 |
| LNG;P0;D;En |
Significance: 0.00501808 |
Significance: 0.033139 |
| wES;LNG;E14.5;D;En |
Significance: 0.00472124 |
Significance: 0.0164539 |
| wES;FaceP;E14.5;D;En |
Significance: 6.97427e-08 |
Significance: 0.00316468 |
| wES;FaceP;E11.5;D;En |
Significance: 9.61692e-18 |
Significance: 4.61911e-08 |
| wES;NRL_Tube;E11.5;D;En |
Significance: 0.00599965 |
Significance: 0.028370299999999998 |
| CRBL_GRNL_Precrsr;A;GEO |
Significance: 0.0505661 |
Significance: 0.0273952 |
| wES.CRNL_NRL_CRST;E10.5;A;GEO |
Significance: 4.7348e-14 |
Significance: 0.00553084 |
| RTN;D1;D;En |
Significance: 0.05111280000000001 |
Significance: 0.0535591 |
| eCortical_NRN;A;GEO |
Significance: 0.0431375 |
Significance: 0.040052 |
| wES;MSDRM;E11.5;D;En |
Significance: 9.38297e-05 |
Significance: 0.020804800000000002 |
| wES;WHL_BRN;E14.5;D;GEO |
Significance: 0.058830999999999994 |
Significance: 0.0461893 |
| 3T3-L1;D;En |
Significance: 1.90716e-17 |
Significance: 0.0345231 |
| KDNY;P0;D;En |
Significance: 0.00022769 |
Significance: 0.039407 |
Unique Cell types only TF-MoDISco finds Twist1: #45
| Cell type | Modisco | Homer |
|---|---|---|
| proB;A;CL |
Significance: 0.05686889999999999 | Absent |
| prePLSM_B;A;GEO |
Significance: 0.0241417 | Absent |
| plasmaB;A;CL |
Significance: 0.0381466 | Absent |
| MCRPHG_BMD.UnStmltd;A;GEO |
Significance: 0.0592695 | Absent |
| NIH3T3;D;En |
Significance: 1.17009e-19 | Absent |
| MTR_NRN_MN1;methyl3;D;En |
Significance: 0.0462225 | Absent |
| npES;3rd_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.059417099999999994 | Absent |
| wES;3rd_Rhmbmr;E8.5;A;GEO |
Significance: 0.00808613 | Absent |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO |
Significance: 0.00107841 | Absent |
| npES;5th_Rhmbmr_delC;E8.5;A;GEO |
Significance: 2.22915e-11 | Absent |
| wES;5th_Rhmbmr;E9.5;A;GEO |
Significance: 0.035518400000000006 | Absent |
| wES;5th_Rhmbmr;E8.5;A;GEO |
Significance: 0.00705909 | Absent |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO |
Significance: 0.0564985 | Absent |
| wES;posterior;1;E9.5;A;GEO |
Significance: 0.000141998 | Absent |
| wES;posterior;2;E9.5;A;GEO |
Significance: 0.00117585 | Absent |
| wES;posterior;1;E8.5;A;GEO |
Significance: 0.00545791 | Absent |
| npES;posterior_delA;E8.5;A;GEO |
Significance: 0.00013022 | Absent |
| npES;posterior_delC;E8.5;A;GEO |
Significance: 0.00039116199999999997 | Absent |
| BCK_SKN;A;GEO |
Significance: 0.057437 | Absent |
| iVIP_NRN;A;GEO |
Significance: 0.022297099999999997 | Absent |
| G_NRN_P;A;GEO |
Significance: 0.055184199999999996 | Absent |
| ES;MEF;A;CL |
Significance: 1.43353e-17 | Absent |
| LNG;A;CL |
Significance: 0.0258278 | Absent |
| Heart;D;En |
Significance: 0.00723974 | Absent |
| pMeg;A;En |
Significance: 0.05985309999999999 | Absent |
| BRN;FRNTL_CRTX;A;CL |
Significance: 0.031653 | Absent |
| BRN;SNSR_MTR_CRTX;A;CL |
Significance: 0.021000799999999997 | Absent |
| NIH3T3_FBROBLST;D;GEO |
Significance: 1.7452300000000002e-23 | Absent |
| LNG;8wks;D;GEO |
Significance: 0.00836789 | Absent |
| WHL_BRN;8wks;D;GEO |
Significance: 0.05159009999999999 | Absent |
| NIH3T3_FBROBLST;D;En |
Significance: 5.998880000000001e-22 | Absent |
| FBRBLST;8wks;D;En |
Significance: 4.9860599999999995e-19 | Absent |
| 3T3-L1;FBRBLST;D;En |
Significance: 6.541419999999999e-18 | Absent |
| LNG;8wks;D;En |
Significance: 0.00128509 | Absent |
| Heart;8wks;D;En |
Significance: 0.0183927 | Absent |
| Adipocyte_3T3-L1D;C57BL6;D8;D;En |
Significance: 3.51179e-19 | Absent |
| wES;RTN;E11.5;D;En |
Significance: 0.028504200000000004 | Absent |
| Muller;P12;D;En |
Significance: 0.056050499999999996 | Absent |
| wES.Fr_BRN;E14.4;D;En |
Significance: 0.0568052 | Absent |
| Fr_BRN;P0;D;En |
Significance: 0.052090599999999994 | Absent |
| wES;Hnd_BRN;E11.5;D;En |
Significance: 0.00497956 | Absent |
| wES;Hnd_BRN;E14.5;D;En |
Significance: 0.0326978 | Absent |
| wES;Md_BRN;E14.4;D;En |
Significance: 0.0385351 | Absent |
| Md_BRN;D;En |
Significance: 0.018080099999999998 | Absent |
| wES;Telenchephalon;D;En |
Significance: 0.0393204 | Absent |
Unique Cell types only Homer finds Twist1: #2
| Cell type | Modisco | Homer |
|---|---|---|
| B;B220_CD43;A;GEO | Absent |
Significance: 0.0485336 |
| ePyramidal_NRN;A;GEO | Absent |
Significance: 0.00878299 |