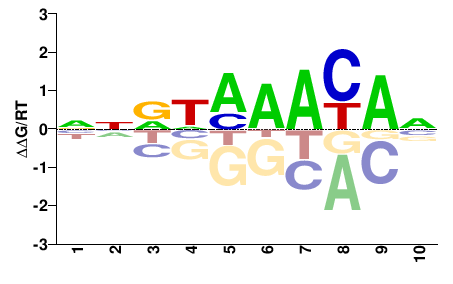
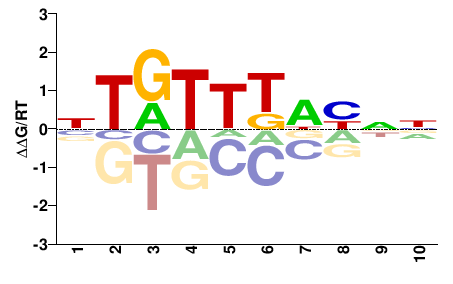
| wES;5th_Rhmbmr;E8.5;A;GEO | Pattern: metacluster_0/pattern_14
Significance: 0.015155200000000002  | Pattern: motif9.motif
Significance: 0.0030464000000000003  |
| npES;5th_Rhmbmr_delA;E8.5;A;GEO | Pattern: metacluster_0/pattern_12
Significance: 0.0152939  | Pattern: motif15.motif
Significance: 0.00921948  |
| LVR;P0;D;En | Pattern: metacluster_1/pattern_14
Significance: 0.00922496  / Pattern: metacluster_1/pattern_18
Significance: 0.0527131  | Pattern: motif16.motif
Significance: 0.00441521  |
| LNG;8wks;D;En | Pattern: metacluster_0/pattern_5
Significance: 0.034968099999999995  | Pattern: motif5.motif
Significance: 0.00494516  |
| LVR;8wks;D;En | Pattern: metacluster_0/pattern_13
Significance: 0.00513532  | Pattern: motif7.motif
Significance: 0.0206908  |
| npES;posterior_delA;E8.5;A;GEO | Pattern: metacluster_0/pattern_10
Significance: 0.00748224  | Pattern: motif15.motif
Significance: 0.037576099999999994  |
| LNG;P0;D;En | Pattern: metacluster_1/pattern_6
Significance: 0.0153268  | Pattern: motif5.motif
Significance: 0.000736386  |
| wES;LNG;E14.5;D;En | Pattern: metacluster_1/pattern_5
Significance: 0.0129666  | Pattern: motif11.motif
Significance: 0.032542  |
| STMCH;P0;D;En | Pattern: metacluster_1/pattern_4
Significance: 0.0198894  | Pattern: motif3.motif
Significance: 0.020981700000000002  |
| LVR;8wks;D;GEO | Pattern: metacluster_1/pattern_4
Significance: 0.014272299999999998  | Pattern: motif4.motif
Significance: 0.0567288  |
| LNG;8wks;D;GEO | Pattern: metacluster_0/pattern_5
Significance: 0.018571200000000003  | Pattern: motif6.motif
Significance: 0.036026499999999996  |
| SML_INTSTN;A;CL | Pattern: metacluster_0/pattern_9
Significance: 0.034536199999999996  | Pattern: motif16.motif
Significance: 0.0378604  |
| ENDRM_ESD;A;GEO | Pattern: metacluster_1/pattern_6
Significance: 0.0124935  / Pattern: metacluster_1/pattern_21
Significance: 0.05411130000000001  | Pattern: motif5.motif
Significance: 0.0202613  |
| wES;3rd_Rhmbmr;E8.5;A;GEO | Pattern: metacluster_0/pattern_13
Significance: 0.0113665  | Pattern: motif15.motif
Significance: 0.00631748  |
| npES;3rd_Rhmbmr_delC;E8.5;A;GEO | Pattern: metacluster_0/pattern_10
Significance: 0.0159776  | Pattern: motif16.motif
Significance: 0.00942607  |