| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | ABF1/SacCer-Promoters/Homer | 1e-222 | -5.126e+02 | 0.0000 | 1469.0 | 35.12% | 3129.6 | 15.12% | motif file (matrix) |
pdf |
| 2 |  | REB1/SacCer-Promoters/Homer | 1e-182 | -4.199e+02 | 0.0000 | 943.0 | 22.54% | 1664.8 | 8.05% | motif file (matrix) |
pdf |
| 3 |  | RLR1?/SacCer-Promoters/Homer | 1e-82 | -1.889e+02 | 0.0000 | 2052.0 | 49.06% | 7150.5 | 34.56% | motif file (matrix) |
pdf |
| 4 |  | SeqBias: CG-repeat | 1e-58 | -1.339e+02 | 0.0000 | 771.0 | 18.43% | 2098.4 | 10.14% | motif file (matrix) |
pdf |
| 5 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-52 | -1.204e+02 | 0.0000 | 566.0 | 13.53% | 1413.3 | 6.83% | motif file (matrix) |
pdf |
| 6 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-45 | -1.049e+02 | 0.0000 | 427.0 | 10.21% | 997.8 | 4.82% | motif file (matrix) |
pdf |
| 7 |  | DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer | 1e-43 | -1.001e+02 | 0.0000 | 1055.0 | 25.22% | 3461.6 | 16.73% | motif file (matrix) |
pdf |
| 8 |  | Cbf1(bHLH)/Yeast-Cbf1-ChIP-Seq(GSE29506)/Homer | 1e-43 | -9.939e+01 | 0.0000 | 409.0 | 9.78% | 960.0 | 4.64% | motif file (matrix) |
pdf |
| 9 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-42 | -9.792e+01 | 0.0000 | 636.0 | 15.20% | 1790.1 | 8.65% | motif file (matrix) |
pdf |
| 10 |  | SFP1/SacCer-Promoters/Homer | 1e-37 | -8.541e+01 | 0.0000 | 619.0 | 14.80% | 1802.5 | 8.71% | motif file (matrix) |
pdf |
| 11 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-36 | -8.453e+01 | 0.0000 | 547.0 | 13.08% | 1532.5 | 7.41% | motif file (matrix) |
pdf |
| 12 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-35 | -8.229e+01 | 0.0000 | 472.0 | 11.28% | 1265.1 | 6.11% | motif file (matrix) |
pdf |
| 13 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-34 | -7.864e+01 | 0.0000 | 380.0 | 9.08% | 952.0 | 4.60% | motif file (matrix) |
pdf |
| 14 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-29 | -6.896e+01 | 0.0000 | 419.0 | 10.02% | 1144.2 | 5.53% | motif file (matrix) |
pdf |
| 15 |  | E-box(bHLH)/Promoter/Homer | 1e-27 | -6.414e+01 | 0.0000 | 132.0 | 3.16% | 210.4 | 1.02% | motif file (matrix) |
pdf |
| 16 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-27 | -6.237e+01 | 0.0000 | 558.0 | 13.34% | 1722.3 | 8.32% | motif file (matrix) |
pdf |
| 17 |  | E-box/Arabidopsis-Promoters/Homer | 1e-26 | -6.107e+01 | 0.0000 | 498.0 | 11.91% | 1495.2 | 7.23% | motif file (matrix) |
pdf |
| 18 |  | TATA-box/SacCer-Promoters/Homer | 1e-24 | -5.755e+01 | 0.0000 | 1918.0 | 45.85% | 7850.6 | 37.94% | motif file (matrix) |
pdf |
| 19 |  | FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer | 1e-24 | -5.669e+01 | 0.0000 | 421.0 | 10.06% | 1228.0 | 5.93% | motif file (matrix) |
pdf |
| 20 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-23 | -5.435e+01 | 0.0000 | 129.0 | 3.08% | 226.1 | 1.09% | motif file (matrix) |
pdf |
| 21 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-21 | -4.992e+01 | 0.0000 | 1274.0 | 30.46% | 4944.3 | 23.89% | motif file (matrix) |
pdf |
| 22 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-21 | -4.956e+01 | 0.0000 | 781.0 | 18.67% | 2761.6 | 13.35% | motif file (matrix) |
pdf |
| 23 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-21 | -4.906e+01 | 0.0000 | 819.0 | 19.58% | 2930.6 | 14.16% | motif file (matrix) |
pdf |
| 24 |  | TOD6?/SacCer-Promoters/Homer | 1e-21 | -4.869e+01 | 0.0000 | 331.0 | 7.91% | 937.5 | 4.53% | motif file (matrix) |
pdf |
| 25 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-18 | -4.211e+01 | 0.0000 | 897.0 | 21.44% | 3353.6 | 16.21% | motif file (matrix) |
pdf |
| 26 |  | NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-17 | -3.930e+01 | 0.0000 | 764.0 | 18.26% | 2804.3 | 13.55% | motif file (matrix) |
pdf |
| 27 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-16 | -3.885e+01 | 0.0000 | 1020.0 | 24.38% | 3946.9 | 19.07% | motif file (matrix) |
pdf |
| 28 |  | KLF10(Zf)/HEK293-KLF10.GFP-ChIP-Seq(GSE58341)/Homer | 1e-14 | -3.327e+01 | 0.0000 | 242.0 | 5.79% | 701.7 | 3.39% | motif file (matrix) |
pdf |
| 29 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-14 | -3.299e+01 | 0.0000 | 532.0 | 12.72% | 1879.6 | 9.08% | motif file (matrix) |
pdf |
| 30 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-13 | -3.103e+01 | 0.0000 | 289.0 | 6.91% | 899.8 | 4.35% | motif file (matrix) |
pdf |
| 31 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo_et_al.)/Homer | 1e-11 | -2.666e+01 | 0.0000 | 992.0 | 23.72% | 4010.9 | 19.38% | motif file (matrix) |
pdf |
| 32 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-11 | -2.598e+01 | 0.0000 | 731.0 | 17.48% | 2839.2 | 13.72% | motif file (matrix) |
pdf |
| 33 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-11 | -2.572e+01 | 0.0000 | 444.0 | 10.61% | 1588.6 | 7.68% | motif file (matrix) |
pdf |
| 34 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-10 | -2.531e+01 | 0.0000 | 299.0 | 7.15% | 988.4 | 4.78% | motif file (matrix) |
pdf |
| 35 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.489e+01 | 0.0000 | 147.0 | 3.51% | 400.9 | 1.94% | motif file (matrix) |
pdf |
| 36 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-10 | -2.442e+01 | 0.0000 | 276.0 | 6.60% | 903.0 | 4.36% | motif file (matrix) |
pdf |
| 37 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.401e+01 | 0.0000 | 315.0 | 7.53% | 1066.0 | 5.15% | motif file (matrix) |
pdf |
| 38 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-10 | -2.386e+01 | 0.0000 | 1326.0 | 31.70% | 5619.2 | 27.16% | motif file (matrix) |
pdf |
| 39 |  | SUT1?/SacCer-Promoters/Homer | 1e-9 | -2.150e+01 | 0.0000 | 2817.0 | 67.34% | 12994.2 | 62.80% | motif file (matrix) |
pdf |
| 40 |  | SeqBias: TA-repeat | 1e-9 | -2.133e+01 | 0.0000 | 3747.0 | 89.58% | 17888.6 | 86.45% | motif file (matrix) |
pdf |
| 41 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-9 | -2.094e+01 | 0.0000 | 122.0 | 2.92% | 332.5 | 1.61% | motif file (matrix) |
pdf |
| 42 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-8 | -1.908e+01 | 0.0000 | 144.0 | 3.44% | 427.0 | 2.06% | motif file (matrix) |
pdf |
| 43 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-8 | -1.901e+01 | 0.0000 | 588.0 | 14.06% | 2311.4 | 11.17% | motif file (matrix) |
pdf |
| 44 |  | CRE(bZIP)/Promoter/Homer | 1e-8 | -1.887e+01 | 0.0000 | 200.0 | 4.78% | 647.5 | 3.13% | motif file (matrix) |
pdf |
| 45 |  | GAGA-repeat/Arabidopsis-Promoters/Homer | 1e-7 | -1.734e+01 | 0.0000 | 488.0 | 11.67% | 1893.5 | 9.15% | motif file (matrix) |
pdf |
| 46 |  | NRF(NRF)/Promoter/Homer | 1e-7 | -1.673e+01 | 0.0000 | 93.0 | 2.22% | 251.9 | 1.22% | motif file (matrix) |
pdf |
| 47 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-7 | -1.662e+01 | 0.0000 | 1555.0 | 37.17% | 6885.7 | 33.28% | motif file (matrix) |
pdf |
| 48 |  | HIF2a(bHLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-6 | -1.565e+01 | 0.0000 | 692.0 | 16.54% | 2843.5 | 13.74% | motif file (matrix) |
pdf |
| 49 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-6 | -1.409e+01 | 0.0000 | 1379.0 | 32.97% | 6110.1 | 29.53% | motif file (matrix) |
pdf |
| 50 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-6 | -1.398e+01 | 0.0000 | 187.0 | 4.47% | 641.1 | 3.10% | motif file (matrix) |
pdf |
| 51 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-5 | -1.367e+01 | 0.0000 | 97.0 | 2.32% | 285.9 | 1.38% | motif file (matrix) |
pdf |
| 52 |  | Pax8(Paired,Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-5 | -1.356e+01 | 0.0000 | 130.0 | 3.11% | 414.2 | 2.00% | motif file (matrix) |
pdf |
| 53 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-5 | -1.238e+01 | 0.0000 | 53.0 | 1.27% | 132.6 | 0.64% | motif file (matrix) |
pdf |
| 54 |  | HIF-1a(bHLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-4 | -1.056e+01 | 0.0002 | 451.0 | 10.78% | 1849.7 | 8.94% | motif file (matrix) |
pdf |
| 55 |  | PAX5(Paired,Homeobox),condensed/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -1.021e+01 | 0.0003 | 79.0 | 1.89% | 241.8 | 1.17% | motif file (matrix) |
pdf |
| 56 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-4 | -9.789e+00 | 0.0004 | 499.0 | 11.93% | 2085.3 | 10.08% | motif file (matrix) |
pdf |
| 57 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-4 | -9.665e+00 | 0.0004 | 40.0 | 0.96% | 100.7 | 0.49% | motif file (matrix) |
pdf |
| 58 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.594e+00 | 0.0005 | 976.0 | 23.33% | 4323.6 | 20.89% | motif file (matrix) |
pdf |
| 59 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-4 | -9.479e+00 | 0.0005 | 275.0 | 6.57% | 1080.0 | 5.22% | motif file (matrix) |
pdf |
| 60 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-3 | -8.435e+00 | 0.0014 | 113.0 | 2.70% | 394.7 | 1.91% | motif file (matrix) |
pdf |
| 61 |  | TATA-box/Drosophila-Promoters/Homer | 1e-3 | -8.245e+00 | 0.0017 | 203.0 | 4.85% | 782.8 | 3.78% | motif file (matrix) |
pdf |
| 62 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-3 | -8.051e+00 | 0.0020 | 117.0 | 2.80% | 415.3 | 2.01% | motif file (matrix) |
pdf |
| 63 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.990e+00 | 0.0021 | 265.0 | 6.34% | 1061.5 | 5.13% | motif file (matrix) |
pdf |
| 64 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-3 | -7.966e+00 | 0.0021 | 63.0 | 1.51% | 196.8 | 0.95% | motif file (matrix) |
pdf |
| 65 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-3 | -7.966e+00 | 0.0021 | 63.0 | 1.51% | 196.8 | 0.95% | motif file (matrix) |
pdf |
| 66 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-3 | -7.739e+00 | 0.0026 | 33.0 | 0.79% | 85.4 | 0.41% | motif file (matrix) |
pdf |
| 67 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.691e+00 | 0.0026 | 589.0 | 14.08% | 2556.2 | 12.35% | motif file (matrix) |
pdf |
| 68 |  | PAX5(Paired,Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-3 | -7.492e+00 | 0.0032 | 200.0 | 4.78% | 781.4 | 3.78% | motif file (matrix) |
pdf |
| 69 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-3 | -7.438e+00 | 0.0033 | 86.0 | 2.06% | 293.6 | 1.42% | motif file (matrix) |
pdf |
| 70 |  | PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-3 | -7.399e+00 | 0.0034 | 481.0 | 11.50% | 2061.0 | 9.96% | motif file (matrix) |
pdf |
| 71 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-3 | -7.380e+00 | 0.0034 | 488.0 | 11.67% | 2094.7 | 10.12% | motif file (matrix) |
pdf |
| 72 |  | Dorsal(RHD)/Embryo-dl-ChIP-Seq(GSE65441)/Homer | 1e-3 | -7.372e+00 | 0.0034 | 200.0 | 4.78% | 783.1 | 3.78% | motif file (matrix) |
pdf |
| 73 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.187e+00 | 0.0040 | 799.0 | 19.10% | 3562.7 | 17.22% | motif file (matrix) |
pdf |
| 74 |  | GEI-11(Myb?)/cElegans-L4-GEI11-ChIP-Seq(modEncode)/Homer | 1e-3 | -7.148e+00 | 0.0041 | 43.0 | 1.03% | 125.0 | 0.60% | motif file (matrix) |
pdf |
| 75 |  | ETS(ETS)/Promoter/Homer | 1e-3 | -6.918e+00 | 0.0051 | 333.0 | 7.96% | 1391.6 | 6.73% | motif file (matrix) |
pdf |
| 76 | 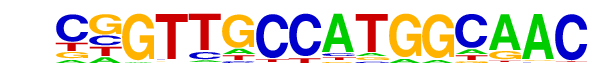 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -6.841e+00 | 0.0054 | 76.0 | 1.82% | 258.8 | 1.25% | motif file (matrix) |
pdf |
| 77 | 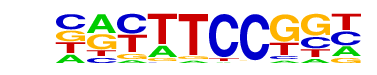 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.763e+00 | 0.0058 | 1265.0 | 30.24% | 5814.7 | 28.10% | motif file (matrix) |
pdf |
| 78 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -6.736e+00 | 0.0059 | 48.0 | 1.15% | 147.5 | 0.71% | motif file (matrix) |
pdf |
| 79 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.682e+00 | 0.0061 | 754.0 | 18.03% | 3367.0 | 16.27% | motif file (matrix) |
pdf |
| 80 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.360e+00 | 0.0084 | 551.0 | 13.17% | 2418.3 | 11.69% | motif file (matrix) |
pdf |
| 81 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -6.339e+00 | 0.0084 | 122.0 | 2.92% | 458.6 | 2.22% | motif file (matrix) |
pdf |
| 82 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-2 | -6.218e+00 | 0.0094 | 177.0 | 4.23% | 701.5 | 3.39% | motif file (matrix) |
pdf |
| 83 |  | Trl(Zf)/S2-GAGAfactor-ChIP-Seq(GSE40646)/Homer | 1e-2 | -6.163e+00 | 0.0098 | 1590.0 | 38.01% | 7422.8 | 35.87% | motif file (matrix) |
pdf |
| 84 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-2 | -6.006e+00 | 0.0114 | 522.0 | 12.48% | 2293.7 | 11.08% | motif file (matrix) |
pdf |
| 85 |  | Tbox:Smad(T-box,MAD)/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-2 | -5.660e+00 | 0.0159 | 98.0 | 2.34% | 365.0 | 1.76% | motif file (matrix) |
pdf |
| 86 |  | TR4(NR),DR1/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-2 | -5.612e+00 | 0.0164 | 46.0 | 1.10% | 148.1 | 0.72% | motif file (matrix) |
pdf |
| 87 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.429e+00 | 0.0195 | 37.0 | 0.88% | 114.2 | 0.55% | motif file (matrix) |
pdf |
| 88 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-2 | -5.258e+00 | 0.0229 | 705.0 | 16.85% | 3186.5 | 15.40% | motif file (matrix) |
pdf |
| 89 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.214e+00 | 0.0237 | 239.0 | 5.71% | 1001.7 | 4.84% | motif file (matrix) |
pdf |
| 90 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -4.697e+00 | 0.0392 | 193.0 | 4.61% | 803.8 | 3.88% | motif file (matrix) |
pdf |
| 91 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-2 | -4.683e+00 | 0.0393 | 1100.0 | 26.30% | 5112.3 | 24.71% | motif file (matrix) |
pdf |





















































































































































































