| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | ABF1/SacCer-Promoters/Homer | 1e-231 | -5.341e+02 | 0.0000 | 1450.0 | 34.76% | 3123.9 | 14.53% | motif file (matrix) |
pdf |
| 2 |  | REB1/SacCer-Promoters/Homer | 1e-190 | -4.382e+02 | 0.0000 | 918.0 | 22.00% | 1610.4 | 7.49% | motif file (matrix) |
pdf |
| 3 |  | RLR1?/SacCer-Promoters/Homer | 1e-79 | -1.825e+02 | 0.0000 | 2003.0 | 48.01% | 7269.3 | 33.80% | motif file (matrix) |
pdf |
| 4 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-68 | -1.568e+02 | 0.0000 | 574.0 | 13.76% | 1341.7 | 6.24% | motif file (matrix) |
pdf |
| 5 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-59 | -1.371e+02 | 0.0000 | 441.0 | 10.57% | 960.9 | 4.47% | motif file (matrix) |
pdf |
| 6 |  | Cbf1(bHLH)/Yeast-Cbf1-ChIP-Seq(GSE29506)/Homer | 1e-56 | -1.293e+02 | 0.0000 | 420.0 | 10.07% | 918.3 | 4.27% | motif file (matrix) |
pdf |
| 7 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-56 | -1.291e+02 | 0.0000 | 648.0 | 15.53% | 1738.0 | 8.08% | motif file (matrix) |
pdf |
| 8 |  | SeqBias: CG-repeat | 1e-55 | -1.273e+02 | 0.0000 | 762.0 | 18.26% | 2189.9 | 10.18% | motif file (matrix) |
pdf |
| 9 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-54 | -1.244e+02 | 0.0000 | 561.0 | 13.45% | 1435.4 | 6.67% | motif file (matrix) |
pdf |
| 10 |  | DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer | 1e-49 | -1.146e+02 | 0.0000 | 1054.0 | 25.26% | 3486.6 | 16.21% | motif file (matrix) |
pdf |
| 11 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-49 | -1.135e+02 | 0.0000 | 484.0 | 11.60% | 1205.5 | 5.61% | motif file (matrix) |
pdf |
| 12 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-46 | -1.072e+02 | 0.0000 | 392.0 | 9.40% | 907.1 | 4.22% | motif file (matrix) |
pdf |
| 13 |  | SFP1/SacCer-Promoters/Homer | 1e-45 | -1.041e+02 | 0.0000 | 642.0 | 15.39% | 1852.6 | 8.61% | motif file (matrix) |
pdf |
| 14 |  | E-box/Arabidopsis-Promoters/Homer | 1e-43 | -9.942e+01 | 0.0000 | 520.0 | 12.46% | 1407.1 | 6.54% | motif file (matrix) |
pdf |
| 15 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-36 | -8.512e+01 | 0.0000 | 564.0 | 13.52% | 1660.6 | 7.72% | motif file (matrix) |
pdf |
| 16 |  | E-box(bHLH)/Promoter/Homer | 1e-34 | -8.015e+01 | 0.0000 | 138.0 | 3.31% | 200.9 | 0.93% | motif file (matrix) |
pdf |
| 17 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-34 | -7.859e+01 | 0.0000 | 413.0 | 9.90% | 1115.7 | 5.19% | motif file (matrix) |
pdf |
| 18 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-33 | -7.824e+01 | 0.0000 | 840.0 | 20.13% | 2859.0 | 13.29% | motif file (matrix) |
pdf |
| 19 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-30 | -6.977e+01 | 0.0000 | 917.0 | 21.98% | 3273.6 | 15.22% | motif file (matrix) |
pdf |
| 20 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-28 | -6.448e+01 | 0.0000 | 1261.0 | 30.23% | 4902.0 | 22.79% | motif file (matrix) |
pdf |
| 21 |  | TATA-box/SacCer-Promoters/Homer | 1e-27 | -6.218e+01 | 0.0000 | 1865.0 | 44.70% | 7849.4 | 36.50% | motif file (matrix) |
pdf |
| 22 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-27 | -6.218e+01 | 0.0000 | 1036.0 | 24.83% | 3888.2 | 18.08% | motif file (matrix) |
pdf |
| 23 |  | TOD6?/SacCer-Promoters/Homer | 1e-25 | -5.789e+01 | 0.0000 | 332.0 | 7.96% | 924.8 | 4.30% | motif file (matrix) |
pdf |
| 24 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-22 | -5.284e+01 | 0.0000 | 756.0 | 18.12% | 2731.8 | 12.70% | motif file (matrix) |
pdf |
| 25 |  | NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-21 | -5.012e+01 | 0.0000 | 754.0 | 18.07% | 2751.4 | 12.79% | motif file (matrix) |
pdf |
| 26 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-20 | -4.632e+01 | 0.0000 | 132.0 | 3.16% | 269.6 | 1.25% | motif file (matrix) |
pdf |
| 27 |  | FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer | 1e-19 | -4.605e+01 | 0.0000 | 393.0 | 9.42% | 1243.2 | 5.78% | motif file (matrix) |
pdf |
| 28 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-19 | -4.392e+01 | 0.0000 | 1327.0 | 31.81% | 5495.1 | 25.55% | motif file (matrix) |
pdf |
| 29 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-18 | -4.246e+01 | 0.0000 | 740.0 | 17.74% | 2775.7 | 12.91% | motif file (matrix) |
pdf |
| 30 |  | KLF10(Zf)/HEK293-KLF10.GFP-ChIP-Seq(GSE58341)/Homer | 1e-17 | -3.965e+01 | 0.0000 | 237.0 | 5.68% | 670.5 | 3.12% | motif file (matrix) |
pdf |
| 31 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-16 | -3.711e+01 | 0.0000 | 322.0 | 7.72% | 1023.9 | 4.76% | motif file (matrix) |
pdf |
| 32 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo_et_al.)/Homer | 1e-13 | -3.223e+01 | 0.0000 | 981.0 | 23.51% | 4031.1 | 18.74% | motif file (matrix) |
pdf |
| 33 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-13 | -3.152e+01 | 0.0000 | 300.0 | 7.19% | 978.8 | 4.55% | motif file (matrix) |
pdf |
| 34 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-13 | -3.144e+01 | 0.0000 | 515.0 | 12.34% | 1901.5 | 8.84% | motif file (matrix) |
pdf |
| 35 |  | SUT1?/SacCer-Promoters/Homer | 1e-12 | -2.842e+01 | 0.0000 | 2789.0 | 66.85% | 13229.0 | 61.51% | motif file (matrix) |
pdf |
| 36 |  | SeqBias: TA-repeat | 1e-11 | -2.701e+01 | 0.0000 | 3707.0 | 88.85% | 18314.7 | 85.16% | motif file (matrix) |
pdf |
| 37 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-11 | -2.571e+01 | 0.0000 | 286.0 | 6.86% | 971.5 | 4.52% | motif file (matrix) |
pdf |
| 38 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-10 | -2.402e+01 | 0.0000 | 60.0 | 1.44% | 116.4 | 0.54% | motif file (matrix) |
pdf |
| 39 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-10 | -2.391e+01 | 0.0000 | 434.0 | 10.40% | 1633.9 | 7.60% | motif file (matrix) |
pdf |
| 40 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-10 | -2.372e+01 | 0.0000 | 142.0 | 3.40% | 405.4 | 1.89% | motif file (matrix) |
pdf |
| 41 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-10 | -2.370e+01 | 0.0000 | 272.0 | 6.52% | 932.0 | 4.33% | motif file (matrix) |
pdf |
| 42 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.336e+01 | 0.0000 | 138.0 | 3.31% | 392.7 | 1.83% | motif file (matrix) |
pdf |
| 43 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-9 | -2.263e+01 | 0.0000 | 122.0 | 2.92% | 336.8 | 1.57% | motif file (matrix) |
pdf |
| 44 |  | CRE(bZIP)/Promoter/Homer | 1e-9 | -2.131e+01 | 0.0000 | 193.0 | 4.63% | 624.9 | 2.91% | motif file (matrix) |
pdf |
| 45 |  | GAGA-repeat/Arabidopsis-Promoters/Homer | 1e-8 | -2.063e+01 | 0.0000 | 475.0 | 11.39% | 1862.9 | 8.66% | motif file (matrix) |
pdf |
| 46 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-6 | -1.610e+01 | 0.0000 | 95.0 | 2.28% | 273.7 | 1.27% | motif file (matrix) |
pdf |
| 47 |  | NRF(NRF)/Promoter/Homer | 1e-6 | -1.536e+01 | 0.0000 | 93.0 | 2.23% | 270.3 | 1.26% | motif file (matrix) |
pdf |
| 48 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-6 | -1.460e+01 | 0.0000 | 121.0 | 2.90% | 386.9 | 1.80% | motif file (matrix) |
pdf |
| 49 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-5 | -1.362e+01 | 0.0000 | 1477.0 | 35.40% | 6873.6 | 31.96% | motif file (matrix) |
pdf |
| 50 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.359e+01 | 0.0000 | 1347.0 | 32.29% | 6223.0 | 28.94% | motif file (matrix) |
pdf |
| 51 |  | HIF2a(bHLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-5 | -1.331e+01 | 0.0000 | 651.0 | 15.60% | 2818.2 | 13.10% | motif file (matrix) |
pdf |
| 52 |  | HIF-1a(bHLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-5 | -1.313e+01 | 0.0000 | 445.0 | 10.67% | 1847.2 | 8.59% | motif file (matrix) |
pdf |
| 53 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.250e+01 | 0.0000 | 949.0 | 22.75% | 4284.6 | 19.92% | motif file (matrix) |
pdf |
| 54 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-5 | -1.207e+01 | 0.0000 | 179.0 | 4.29% | 655.5 | 3.05% | motif file (matrix) |
pdf |
| 55 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-5 | -1.202e+01 | 0.0000 | 114.0 | 2.73% | 379.3 | 1.76% | motif file (matrix) |
pdf |
| 56 |  | Pax8(Paired,Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-5 | -1.165e+01 | 0.0001 | 124.0 | 2.97% | 424.5 | 1.97% | motif file (matrix) |
pdf |
| 57 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-4 | -1.083e+01 | 0.0001 | 523.0 | 12.54% | 2263.3 | 10.52% | motif file (matrix) |
pdf |
| 58 |  | ETS(ETS)/Promoter/Homer | 1e-4 | -1.080e+01 | 0.0001 | 322.0 | 7.72% | 1318.1 | 6.13% | motif file (matrix) |
pdf |
| 59 |  | TR4(NR),DR1/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-4 | -1.027e+01 | 0.0002 | 53.0 | 1.27% | 149.6 | 0.70% | motif file (matrix) |
pdf |
| 60 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-4 | -9.959e+00 | 0.0003 | 75.0 | 1.80% | 237.0 | 1.10% | motif file (matrix) |
pdf |
| 61 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.898e+00 | 0.0003 | 767.0 | 18.38% | 3467.7 | 16.12% | motif file (matrix) |
pdf |
| 62 |  | PAX5(Paired,Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-4 | -9.594e+00 | 0.0004 | 191.0 | 4.58% | 740.5 | 3.44% | motif file (matrix) |
pdf |
| 63 |  | Dorsal(RHD)/Embryo-dl-ChIP-Seq(GSE65441)/Homer | 1e-3 | -8.984e+00 | 0.0008 | 197.0 | 4.72% | 776.3 | 3.61% | motif file (matrix) |
pdf |
| 64 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -8.889e+00 | 0.0008 | 525.0 | 12.58% | 2320.4 | 10.79% | motif file (matrix) |
pdf |
| 65 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.686e+00 | 0.0010 | 723.0 | 17.33% | 3288.7 | 15.29% | motif file (matrix) |
pdf |
| 66 |  | PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-3 | -8.623e+00 | 0.0011 | 453.0 | 10.86% | 1981.5 | 9.21% | motif file (matrix) |
pdf |
| 67 | 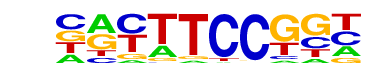 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.463e+00 | 0.0012 | 1219.0 | 29.22% | 5757.8 | 26.77% | motif file (matrix) |
pdf |
| 68 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -8.391e+00 | 0.0013 | 40.0 | 0.96% | 111.7 | 0.52% | motif file (matrix) |
pdf |
| 69 |  | GATA:SCL(Zf,bHLH)/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-3 | -7.359e+00 | 0.0036 | 104.0 | 2.49% | 385.0 | 1.79% | motif file (matrix) |
pdf |
| 70 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-3 | -7.128e+00 | 0.0044 | 681.0 | 16.32% | 3132.7 | 14.57% | motif file (matrix) |
pdf |
| 71 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-3 | -6.910e+00 | 0.0054 | 159.0 | 3.81% | 636.6 | 2.96% | motif file (matrix) |
pdf |
| 72 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-2 | -6.832e+00 | 0.0058 | 455.0 | 10.91% | 2038.5 | 9.48% | motif file (matrix) |
pdf |
| 73 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-2 | -6.795e+00 | 0.0059 | 109.0 | 2.61% | 413.8 | 1.92% | motif file (matrix) |
pdf |
| 74 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-2 | -6.460e+00 | 0.0082 | 256.0 | 6.14% | 1095.9 | 5.10% | motif file (matrix) |
pdf |
| 75 |  | Zic(Zf)/Cerebellum-ZIC1.2-ChIP-Seq(GSE60731)/Homer | 1e-2 | -6.453e+00 | 0.0082 | 357.0 | 8.56% | 1576.3 | 7.33% | motif file (matrix) |
pdf |
| 76 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-2 | -6.211e+00 | 0.0102 | 464.0 | 11.12% | 2100.1 | 9.76% | motif file (matrix) |
pdf |
| 77 |  | NFkB-p50,p52(RHD)/Monocyte-p50-ChIP-Chip(Schreiber_et_al.)/Homer | 1e-2 | -5.810e+00 | 0.0151 | 34.0 | 0.81% | 104.4 | 0.49% | motif file (matrix) |
pdf |
| 78 |  | Trl(Zf)/S2-GAGAfactor-ChIP-Seq(GSE40646)/Homer | 1e-2 | -5.807e+00 | 0.0151 | 1513.0 | 36.27% | 7361.3 | 34.23% | motif file (matrix) |
pdf |
| 79 |  | GEI-11(Myb?)/cElegans-L4-GEI11-ChIP-Seq(modEncode)/Homer | 1e-2 | -5.806e+00 | 0.0151 | 39.0 | 0.93% | 124.1 | 0.58% | motif file (matrix) |
pdf |
| 80 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-2 | -5.495e+00 | 0.0199 | 669.0 | 16.04% | 3132.2 | 14.56% | motif file (matrix) |
pdf |
| 81 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-2 | -5.478e+00 | 0.0200 | 73.0 | 1.75% | 271.5 | 1.26% | motif file (matrix) |
pdf |
| 82 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-2 | -5.415e+00 | 0.0210 | 29.0 | 0.70% | 87.1 | 0.40% | motif file (matrix) |
pdf |
| 83 |  | TATA-box/Drosophila-Promoters/Homer | 1e-2 | -5.260e+00 | 0.0242 | 187.0 | 4.48% | 796.9 | 3.71% | motif file (matrix) |
pdf |
| 84 |  | ARE(NR)/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.135e+00 | 0.0271 | 120.0 | 2.88% | 487.3 | 2.27% | motif file (matrix) |
pdf |
| 85 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-2 | -4.762e+00 | 0.0389 | 294.0 | 7.05% | 1319.6 | 6.14% | motif file (matrix) |
pdf |
| 86 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.721e+00 | 0.0401 | 232.0 | 5.56% | 1022.5 | 4.75% | motif file (matrix) |
pdf |
| 87 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-2 | -4.706e+00 | 0.0402 | 106.0 | 2.54% | 431.0 | 2.00% | motif file (matrix) |
pdf |
| 88 |  | GFY(?)/Promoter/Homer | 1e-2 | -4.656e+00 | 0.0418 | 36.0 | 0.86% | 121.8 | 0.57% | motif file (matrix) |
pdf |















































































































































































