| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | ABF1/SacCer-Promoters/Homer | 1e-219 | -5.050e+02 | 0.0000 | 1491.0 | 35.17% | 3139.5 | 15.38% | motif file (matrix) |
pdf |
| 2 |  | REB1/SacCer-Promoters/Homer | 1e-185 | -4.271e+02 | 0.0000 | 941.0 | 22.20% | 1594.9 | 7.82% | motif file (matrix) |
pdf |
| 3 |  | RLR1?/SacCer-Promoters/Homer | 1e-70 | -1.632e+02 | 0.0000 | 1948.0 | 45.95% | 6679.6 | 32.73% | motif file (matrix) |
pdf |
| 4 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-54 | -1.251e+02 | 0.0000 | 587.0 | 13.85% | 1426.5 | 6.99% | motif file (matrix) |
pdf |
| 5 |  | SFP1/SacCer-Promoters/Homer | 1e-53 | -1.231e+02 | 0.0000 | 644.0 | 15.19% | 1637.0 | 8.02% | motif file (matrix) |
pdf |
| 6 |  | SeqBias: CG-repeat | 1e-50 | -1.155e+02 | 0.0000 | 778.0 | 18.35% | 2169.3 | 10.63% | motif file (matrix) |
pdf |
| 7 |  | DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer | 1e-42 | -9.728e+01 | 0.0000 | 1074.0 | 25.34% | 3466.6 | 16.99% | motif file (matrix) |
pdf |
| 8 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-40 | -9.333e+01 | 0.0000 | 433.0 | 10.21% | 1040.0 | 5.10% | motif file (matrix) |
pdf |
| 9 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-40 | -9.306e+01 | 0.0000 | 672.0 | 15.85% | 1905.9 | 9.34% | motif file (matrix) |
pdf |
| 10 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-39 | -9.209e+01 | 0.0000 | 563.0 | 13.28% | 1506.2 | 7.38% | motif file (matrix) |
pdf |
| 11 |  | Cbf1(bHLH)/Yeast-Cbf1-ChIP-Seq(GSE29506)/Homer | 1e-36 | -8.519e+01 | 0.0000 | 404.0 | 9.53% | 978.5 | 4.79% | motif file (matrix) |
pdf |
| 12 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-36 | -8.508e+01 | 0.0000 | 398.0 | 9.39% | 958.1 | 4.69% | motif file (matrix) |
pdf |
| 13 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-33 | -7.735e+01 | 0.0000 | 476.0 | 11.23% | 1272.1 | 6.23% | motif file (matrix) |
pdf |
| 14 |  | TOD6?/SacCer-Promoters/Homer | 1e-32 | -7.445e+01 | 0.0000 | 373.0 | 8.80% | 922.8 | 4.52% | motif file (matrix) |
pdf |
| 15 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-31 | -7.364e+01 | 0.0000 | 603.0 | 14.23% | 1770.0 | 8.67% | motif file (matrix) |
pdf |
| 16 |  | E-box/Arabidopsis-Promoters/Homer | 1e-27 | -6.329e+01 | 0.0000 | 525.0 | 12.38% | 1542.9 | 7.56% | motif file (matrix) |
pdf |
| 17 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-26 | -6.034e+01 | 0.0000 | 1345.0 | 31.73% | 4991.7 | 24.46% | motif file (matrix) |
pdf |
| 18 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-24 | -5.685e+01 | 0.0000 | 875.0 | 20.64% | 3004.6 | 14.72% | motif file (matrix) |
pdf |
| 19 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-24 | -5.637e+01 | 0.0000 | 428.0 | 10.10% | 1222.8 | 5.99% | motif file (matrix) |
pdf |
| 20 |  | E-box(bHLH)/Promoter/Homer | 1e-23 | -5.461e+01 | 0.0000 | 130.0 | 3.07% | 223.0 | 1.09% | motif file (matrix) |
pdf |
| 21 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-22 | -5.273e+01 | 0.0000 | 964.0 | 22.74% | 3424.4 | 16.78% | motif file (matrix) |
pdf |
| 22 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-22 | -5.198e+01 | 0.0000 | 1081.0 | 25.50% | 3936.6 | 19.29% | motif file (matrix) |
pdf |
| 23 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-21 | -4.899e+01 | 0.0000 | 782.0 | 18.45% | 2696.2 | 13.21% | motif file (matrix) |
pdf |
| 24 |  | FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer | 1e-21 | -4.851e+01 | 0.0000 | 408.0 | 9.62% | 1200.2 | 5.88% | motif file (matrix) |
pdf |
| 25 |  | TATA-box/SacCer-Promoters/Homer | 1e-20 | -4.833e+01 | 0.0000 | 1807.0 | 42.63% | 7253.8 | 35.54% | motif file (matrix) |
pdf |
| 26 |  | NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-20 | -4.808e+01 | 0.0000 | 810.0 | 19.11% | 2823.9 | 13.84% | motif file (matrix) |
pdf |
| 27 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-19 | -4.479e+01 | 0.0000 | 139.0 | 3.28% | 277.8 | 1.36% | motif file (matrix) |
pdf |
| 28 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo_et_al.)/Homer | 1e-15 | -3.570e+01 | 0.0000 | 1044.0 | 24.63% | 3989.0 | 19.55% | motif file (matrix) |
pdf |
| 29 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-14 | -3.275e+01 | 0.0000 | 1394.0 | 32.89% | 5609.1 | 27.49% | motif file (matrix) |
pdf |
| 30 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-12 | -2.796e+01 | 0.0000 | 763.0 | 18.00% | 2872.6 | 14.08% | motif file (matrix) |
pdf |
| 31 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-12 | -2.773e+01 | 0.0000 | 314.0 | 7.41% | 999.5 | 4.90% | motif file (matrix) |
pdf |
| 32 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-11 | -2.614e+01 | 0.0000 | 529.0 | 12.48% | 1895.6 | 9.29% | motif file (matrix) |
pdf |
| 33 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-10 | -2.398e+01 | 0.0000 | 288.0 | 6.79% | 930.5 | 4.56% | motif file (matrix) |
pdf |
| 34 |  | KLF10(Zf)/HEK293-KLF10.GFP-ChIP-Seq(GSE58341)/Homer | 1e-10 | -2.309e+01 | 0.0000 | 235.0 | 5.54% | 730.9 | 3.58% | motif file (matrix) |
pdf |
| 35 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-9 | -2.194e+01 | 0.0000 | 332.0 | 7.83% | 1127.4 | 5.52% | motif file (matrix) |
pdf |
| 36 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-9 | -2.174e+01 | 0.0000 | 150.0 | 3.54% | 419.3 | 2.05% | motif file (matrix) |
pdf |
| 37 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-9 | -2.085e+01 | 0.0000 | 308.0 | 7.27% | 1041.1 | 5.10% | motif file (matrix) |
pdf |
| 38 |  | SeqBias: TA-repeat | 1e-8 | -2.007e+01 | 0.0000 | 3696.0 | 87.19% | 17132.3 | 83.95% | motif file (matrix) |
pdf |
| 39 |  | SUT1?/SacCer-Promoters/Homer | 1e-8 | -1.998e+01 | 0.0000 | 2936.0 | 69.26% | 13262.2 | 64.99% | motif file (matrix) |
pdf |
| 40 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-8 | -1.878e+01 | 0.0000 | 154.0 | 3.63% | 454.6 | 2.23% | motif file (matrix) |
pdf |
| 41 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-7 | -1.809e+01 | 0.0000 | 1604.0 | 37.84% | 6889.3 | 33.76% | motif file (matrix) |
pdf |
| 42 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.787e+01 | 0.0000 | 1385.0 | 32.67% | 5873.8 | 28.78% | motif file (matrix) |
pdf |
| 43 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.755e+01 | 0.0000 | 56.0 | 1.32% | 117.0 | 0.57% | motif file (matrix) |
pdf |
| 44 |  | GAGA-repeat/Arabidopsis-Promoters/Homer | 1e-6 | -1.569e+01 | 0.0000 | 505.0 | 11.91% | 1942.2 | 9.52% | motif file (matrix) |
pdf |
| 45 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-6 | -1.569e+01 | 0.0000 | 444.0 | 10.47% | 1678.8 | 8.23% | motif file (matrix) |
pdf |
| 46 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-6 | -1.531e+01 | 0.0000 | 122.0 | 2.88% | 359.8 | 1.76% | motif file (matrix) |
pdf |
| 47 |  | HIF2a(bHLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-5 | -1.260e+01 | 0.0000 | 715.0 | 16.87% | 2934.0 | 14.38% | motif file (matrix) |
pdf |
| 48 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-5 | -1.231e+01 | 0.0000 | 101.0 | 2.38% | 302.7 | 1.48% | motif file (matrix) |
pdf |
| 49 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-5 | -1.205e+01 | 0.0000 | 181.0 | 4.27% | 620.2 | 3.04% | motif file (matrix) |
pdf |
| 50 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-5 | -1.180e+01 | 0.0001 | 120.0 | 2.83% | 379.8 | 1.86% | motif file (matrix) |
pdf |
| 51 |  | HIF-1a(bHLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-4 | -1.132e+01 | 0.0001 | 458.0 | 10.80% | 1815.1 | 8.89% | motif file (matrix) |
pdf |
| 52 |  | TATA-box/Drosophila-Promoters/Homer | 1e-4 | -1.063e+01 | 0.0002 | 200.0 | 4.72% | 715.3 | 3.51% | motif file (matrix) |
pdf |
| 53 |  | Pax8(Paired,Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-4 | -1.035e+01 | 0.0002 | 132.0 | 3.11% | 440.8 | 2.16% | motif file (matrix) |
pdf |
| 54 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-4 | -1.033e+01 | 0.0002 | 561.0 | 13.23% | 2294.0 | 11.24% | motif file (matrix) |
pdf |
| 55 |  | CRE(bZIP)/Promoter/Homer | 1e-4 | -1.032e+01 | 0.0002 | 194.0 | 4.58% | 694.5 | 3.40% | motif file (matrix) |
pdf |
| 56 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-4 | -1.015e+01 | 0.0003 | 129.0 | 3.04% | 430.2 | 2.11% | motif file (matrix) |
pdf |
| 57 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-4 | -9.945e+00 | 0.0003 | 45.0 | 1.06% | 113.7 | 0.56% | motif file (matrix) |
pdf |
| 58 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-4 | -9.940e+00 | 0.0003 | 713.0 | 16.82% | 2990.8 | 14.66% | motif file (matrix) |
pdf |
| 59 |  | PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-4 | -9.242e+00 | 0.0006 | 490.0 | 11.56% | 2001.8 | 9.81% | motif file (matrix) |
pdf |
| 60 |  | Dorsal(RHD)/Embryo-dl-ChIP-Seq(GSE65441)/Homer | 1e-4 | -9.235e+00 | 0.0006 | 210.0 | 4.95% | 776.1 | 3.80% | motif file (matrix) |
pdf |
| 61 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-3 | -8.938e+00 | 0.0008 | 90.0 | 2.12% | 288.0 | 1.41% | motif file (matrix) |
pdf |
| 62 |  | NRF(NRF)/Promoter/Homer | 1e-3 | -8.896e+00 | 0.0009 | 95.0 | 2.24% | 307.9 | 1.51% | motif file (matrix) |
pdf |
| 63 | 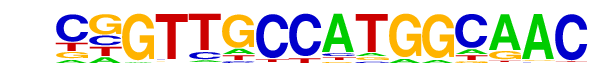 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-3 | -8.854e+00 | 0.0009 | 79.0 | 1.86% | 245.7 | 1.20% | motif file (matrix) |
pdf |
| 64 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.537e+00 | 0.0012 | 1001.0 | 23.61% | 4356.7 | 21.35% | motif file (matrix) |
pdf |
| 65 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-2 | -6.829e+00 | 0.0064 | 34.0 | 0.80% | 91.4 | 0.45% | motif file (matrix) |
pdf |
| 66 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-2 | -6.824e+00 | 0.0064 | 498.0 | 11.75% | 2098.1 | 10.28% | motif file (matrix) |
pdf |
| 67 |  | ERE(NR),IR3/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.629e+00 | 0.0076 | 123.0 | 2.90% | 446.2 | 2.19% | motif file (matrix) |
pdf |
| 68 |  | EFL-1(E2F)/cElegans-L1-EFL1-ChIP-Seq(modEncode)/Homer | 1e-2 | -6.405e+00 | 0.0094 | 23.0 | 0.54% | 55.4 | 0.27% | motif file (matrix) |
pdf |
| 69 |  | GATA:SCL(Zf,bHLH)/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-2 | -6.171e+00 | 0.0117 | 106.0 | 2.50% | 381.2 | 1.87% | motif file (matrix) |
pdf |
| 70 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-2 | -6.159e+00 | 0.0117 | 266.0 | 6.28% | 1073.8 | 5.26% | motif file (matrix) |
pdf |
| 71 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.034e+00 | 0.0131 | 262.0 | 6.18% | 1058.5 | 5.19% | motif file (matrix) |
pdf |
| 72 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-2 | -5.937e+00 | 0.0140 | 69.0 | 1.63% | 232.3 | 1.14% | motif file (matrix) |
pdf |
| 73 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-2 | -5.937e+00 | 0.0140 | 69.0 | 1.63% | 232.3 | 1.14% | motif file (matrix) |
pdf |
| 74 |  | HEB(bHLH)/mES-Heb-ChIP-Seq(GSE53233)/Homer | 1e-2 | -5.918e+00 | 0.0141 | 904.0 | 21.33% | 4000.8 | 19.60% | motif file (matrix) |
pdf |
| 75 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.860e+00 | 0.0147 | 809.0 | 19.08% | 3560.6 | 17.45% | motif file (matrix) |
pdf |
| 76 |  | p63(p53)/Keratinocyte-p63-ChIP-Seq(GSE17611)/Homer | 1e-2 | -5.653e+00 | 0.0179 | 201.0 | 4.74% | 797.9 | 3.91% | motif file (matrix) |
pdf |
| 77 |  | Zic(Zf)/Cerebellum-ZIC1.2-ChIP-Seq(GSE60731)/Homer | 1e-2 | -5.610e+00 | 0.0184 | 395.0 | 9.32% | 1665.1 | 8.16% | motif file (matrix) |
pdf |
| 78 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-2 | -5.465e+00 | 0.0210 | 178.0 | 4.20% | 700.1 | 3.43% | motif file (matrix) |
pdf |
| 79 |  | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.283e+00 | 0.0249 | 1289.0 | 30.41% | 5837.3 | 28.60% | motif file (matrix) |
pdf |
| 80 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-2 | -5.167e+00 | 0.0276 | 126.0 | 2.97% | 480.0 | 2.35% | motif file (matrix) |
pdf |
| 81 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.145e+00 | 0.0278 | 255.0 | 6.02% | 1047.5 | 5.13% | motif file (matrix) |
pdf |
| 82 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -5.062e+00 | 0.0299 | 201.0 | 4.74% | 809.9 | 3.97% | motif file (matrix) |
pdf |
| 83 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-2 | -5.017e+00 | 0.0309 | 302.0 | 7.12% | 1261.0 | 6.18% | motif file (matrix) |
pdf |
| 84 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-2 | -4.921e+00 | 0.0336 | 475.0 | 11.21% | 2051.6 | 10.05% | motif file (matrix) |
pdf |
| 85 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.873e+00 | 0.0348 | 583.0 | 13.75% | 2550.8 | 12.50% | motif file (matrix) |
pdf |
| 86 |  | ETS(ETS)/Promoter/Homer | 1e-2 | -4.788e+00 | 0.0375 | 321.0 | 7.57% | 1353.2 | 6.63% | motif file (matrix) |
pdf |
| 87 |  | Trl(Zf)/S2-GAGAfactor-ChIP-Seq(GSE40646)/Homer | 1e-2 | -4.747e+00 | 0.0386 | 1597.0 | 37.67% | 7327.5 | 35.91% | motif file (matrix) |
pdf |
| 88 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -4.713e+00 | 0.0395 | 42.0 | 0.99% | 136.4 | 0.67% | motif file (matrix) |
pdf |
| 89 |  | Tbox:Smad(T-box,MAD)/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer | 1e-2 | -4.703e+00 | 0.0395 | 100.0 | 2.36% | 376.0 | 1.84% | motif file (matrix) |
pdf |
| 90 |  | Srebp2(bHLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.612e+00 | 0.0427 | 71.0 | 1.67% | 255.3 | 1.25% | motif file (matrix) |
pdf |
| 91 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-2 | -4.610e+00 | 0.0427 | 750.0 | 17.69% | 3336.3 | 16.35% | motif file (matrix) |
pdf |





















































































































































































