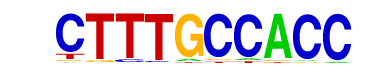
| p-value: | 1e-7 |
| log p-value: | -1.652e+01 |
| Information Content per bp: | 1.815 |
| Number of Target Sequences with motif | 26.0 |
| Percentage of Target Sequences with motif | 4.27% |
| Number of Background Sequences with motif | 557.8 |
| Percentage of Background Sequences with motif | 1.22% |
| Average Position of motif in Targets | 87.8 +/- 52.3bp |
| Average Position of motif in Background | 103.5 +/- 73.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.08 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
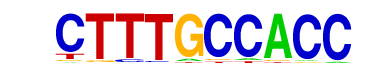
RPN4/RPN4_H2O2Lo/[](Harbison)/Yeast
| Match Rank: | 1 |
| Score: | 0.96
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTTTGCCACC
-TTTGCCACC |
|


|
|
RPN4(MacIsaac)/Yeast
| Match Rank: | 2 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTTTGCCACC-
TTTTGCCACCG |
|


|
|
RPN4/MA0373.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CTTTGCCACC
---CGCCACC |
|

|
|
PB0029.1_Hic1_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTTTGCCACC-----
ACTATGCCAACCTACC |
|


|
|
Hic1/MA0739.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.67
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CTTTGCCACC-
--ATGCCAACC |
|


|
|
slbo/dmmpmm(Bigfoot)/fly
| Match Rank: | 6 |
| Score: | 0.66
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTTTGCCACC-
-TTTGCAATCA |
|


|
|
TBX2/MA0688.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTTTGCCACC--
-TTTCACACCTN |
|

|
|
NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 8 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTTTGCCACC
-CTTGGCAA- |
|

|
|
NFATC2/MA0152.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTTTGCCACC
-TTTTCCA-- |
|

|
|
Six4/dmmpmm(Noyes_hd)/fly
| Match Rank: | 10 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTTTGCCACC
ATTTGATACC |
|


|
|