| p-value: | 1e-20 |
| log p-value: | -4.713e+01 |
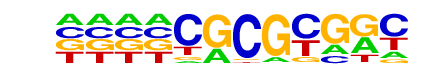
| Information Content per bp: | 1.577 |
| Number of Target Sequences with motif | 857.0 |
| Percentage of Target Sequences with motif | 53.16% |
| Number of Background Sequences with motif | 18537.7 |
| Percentage of Background Sequences with motif | 41.53% |
| Average Position of motif in Targets | 100.8 +/- 52.9bp |
| Average Position of motif in Background | 100.1 +/- 81.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.53 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
RSC3/MA0374.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.89
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGCGCGRC
CGCGCGG- |
|


|
|
RSC30/MA0375.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.85
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CGCGCGRC
CGCGCGCG-- |
|


|
|
Tcfl5/MA0632.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.84
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CGCGCGRC
GGCACGTGCC |
|


|
|
MBP1/MA0329.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.81
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGCGCGRC
NNCGCGT--- |
|


|
|
Hes1/MA1099.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.80
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGCGCGRC
NNCGCGTGNN |
|


|
|
SUT1/MA0399.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGCGCGRC
CGCGGGG- |
|


|
|
DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer
| Match Rank: | 7 |
| Score: | 0.79
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CGCGCGRC
TAGCGCGC--- |
|


|
|
SUT1(MacIsaac)/Yeast
| Match Rank: | 8 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGCGCGRC
CGCGGGG- |
|


|
|
Ahr::Arnt/MA0006.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.76
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGCGCGRC
TGCGTG-- |
|


|
|
FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer
| Match Rank: | 10 |
| Score: | 0.76
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CGCGCGRC
NAVGCGCGTGDD |
|

|
|





















