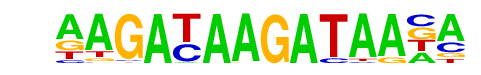| p-value: | 1e-10 |
| log p-value: | -2.476e+01 |
| Information Content per bp: | 1.833 |
| Number of Target Sequences with motif | 13.0 |
| Percentage of Target Sequences with motif | 0.81% |
| Number of Background Sequences with motif | 25.8 |
| Percentage of Background Sequences with motif | 0.06% |
| Average Position of motif in Targets | 101.4 +/- 54.6bp |
| Average Position of motif in Background | 100.4 +/- 54.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ARR10(GARP)/Arabidopsis thaliana/AthaMap
| Match Rank: | 1 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GAGAAGATCATC
GCGAAGATCCGC |
|


|
|
ZmHOX2a(2)(HD-HOX)/Zea mays/AthaMap
| Match Rank: | 2 |
| Score: | 0.62
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GAGAAGATCATC
---CAGATCA-- |
|


|
|
PH0037.1_Hdx/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAGAAGATCATC---
AAGGCGAAATCATCGCA |
|


|
|
Mecom/MA0029.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAGAAGATCATC
AAGATAAGATAACA |
|

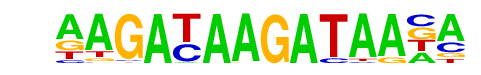
|
|
YPR013C/MA0434.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GAGAAGATCATC
-TGTAGATCA-- |
|


|
|
ECM23/MA0293.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GAGAAGATCATC-
--AAAGATCTAAA |
|


|
|
JUND(var.2)/MA0492.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GAGAAGATCATC---
NATGACATCATCNNN |
|


|
|
BATF3/MA0835.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GAGAAGATCATC-
TGATGACGTCATCA |
|


|
|
JUN/MA0488.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GAGAAGATCATC--
-ATGACATCATCNN |
|


|
|
TOD6/MA0350.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAGAAGATCATC-------
AGGCACAGCTCATCGCGTTTT |
|


|
|