

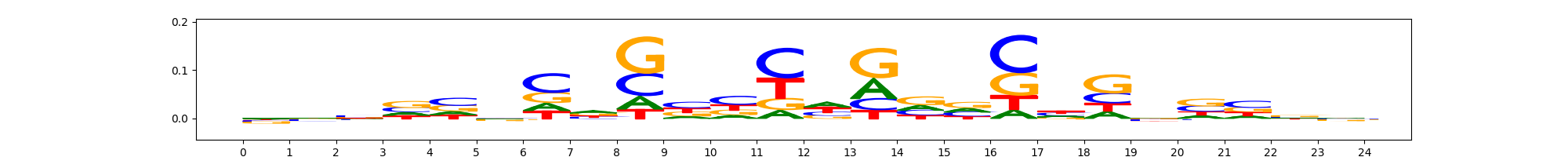
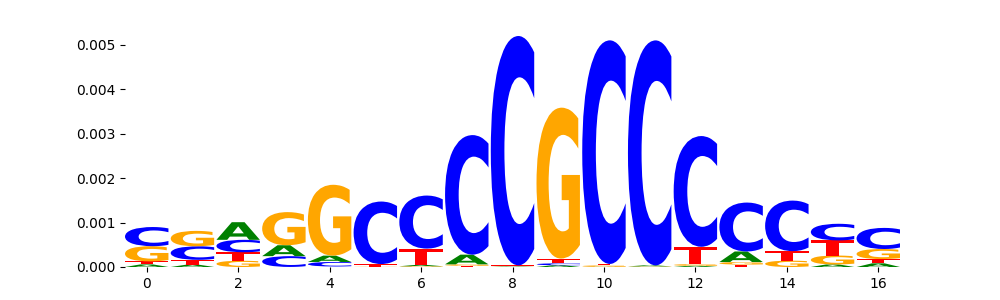
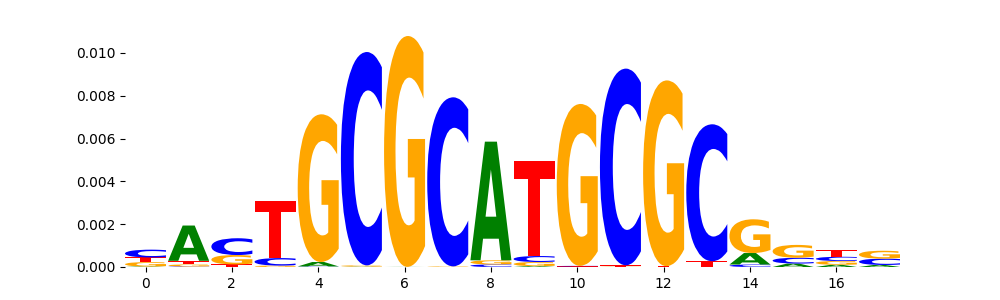
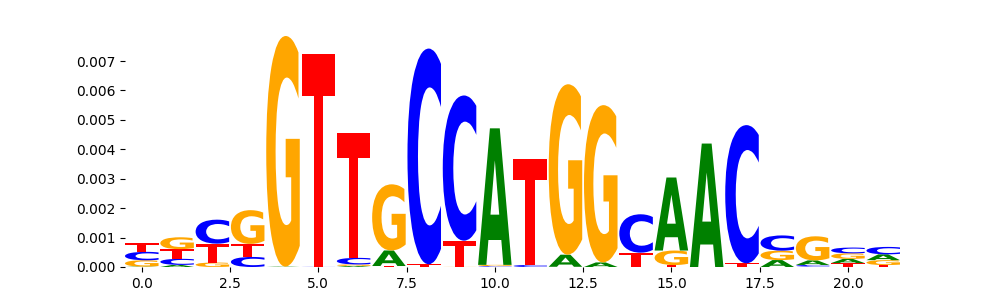
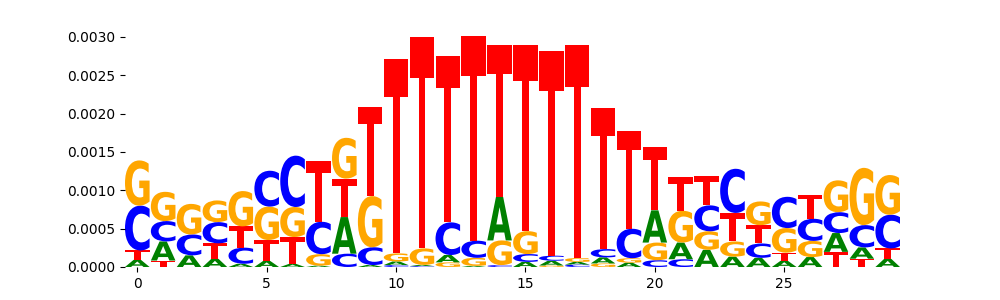
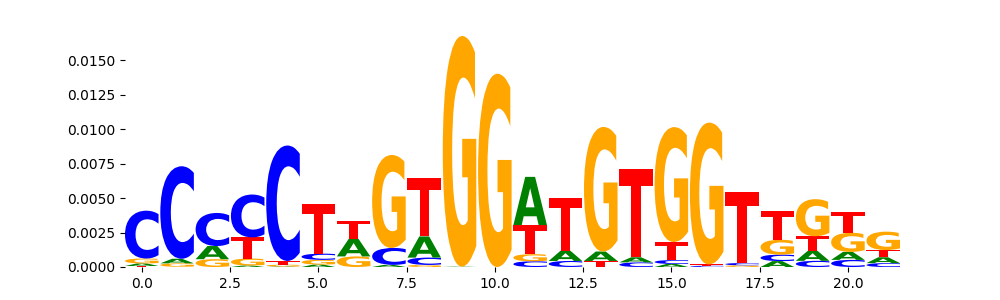
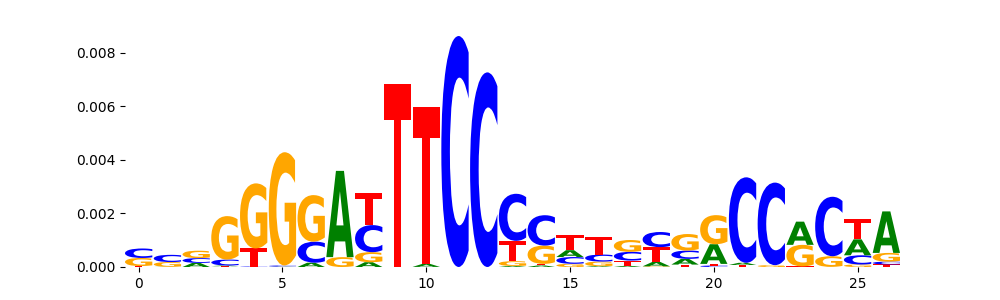
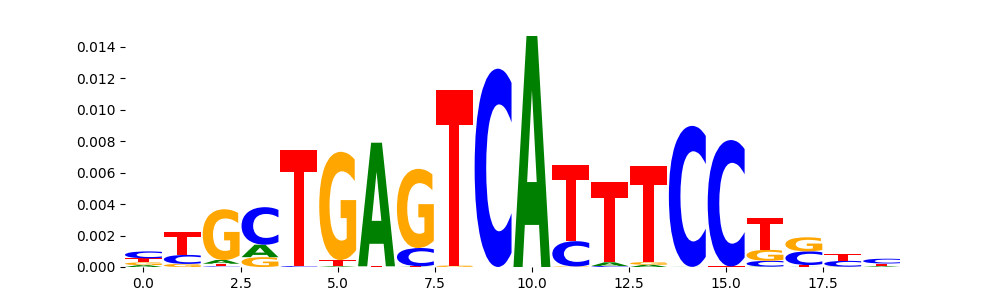
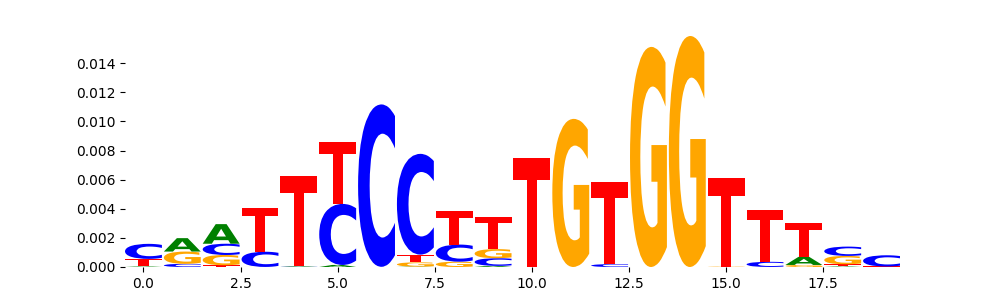
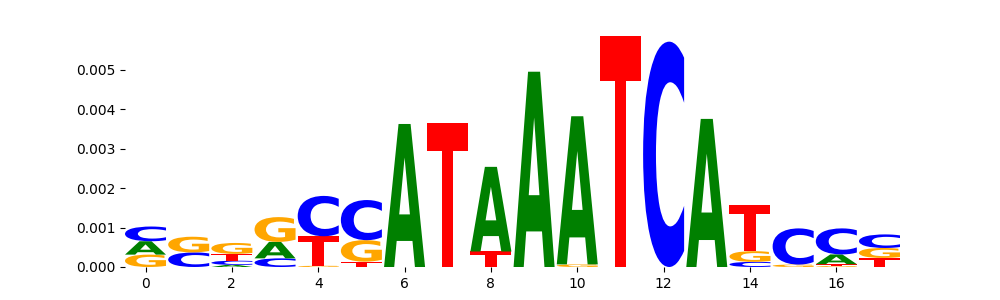
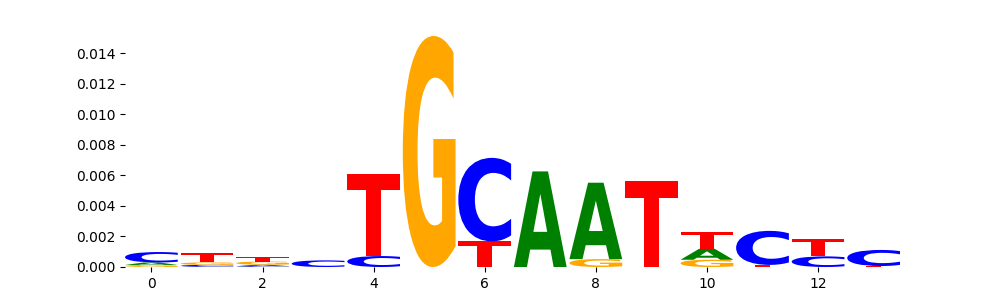
The image below should look closely like a Tn5 or DNase bias enzyme motif.
Counts Metrics: The pearsonr in peaks should be greater than -0.3 (otherwise the bias model could potentially be capturing AT bias). MSE (Mean Squared Error) will be high in peaks.
Profile Metrics: Median JSD (Jensen Shannon Divergence between observed and predicted) lower the better. Median norm JSD is median of the min-max normalized JSD where min JSD is the worst case JSD i.e JSD of observed with uniform profile and max JSD is the best case JSD i.e 0. Median norm JSD is higher the better. Both JSD and median norm JSD are sensitive to read-depth. Higher read-depth results in better metrics.
What to do if your pearsonr in peaks is less than -0.3? In the range of -0.3 to -0.5 please be wary of your chrombpnet_wo_bias.h5 TFModisco results showing lots of GC rich motifs (> 3 in the top-10). If this is not the case you can continue using the chrombpnet_wo_bias.h5. If you end up seeing a lot of GC rich motifs it is likely that bias model has learnt a different GC distribution than your GC-content in peaks. If you are transferring a bias model from a different sample you can consider using a different bias model or training a bias model for this sample. If you have trained a bias model for this sample and encounter this you might have to increase the bias_threshold_factor argument input to the chrombpnet bias pipeline or chrombpnet bias train command used in training the bias model and retrain a new bias model. For more intuition about this argument refer to the FAQ section in wiki. If the value is less than -0.5 the pipline will automatically throw an error.
| peaks.pearsonr | peaks.mse | |
|---|---|---|
| counts_metrics | 0.188456 | 15.097933 |
| peaks.median_jsd | peaks.median_norm_jsd | |
|---|---|---|
| profile_metrics | 0.456584 | 0.303574 |
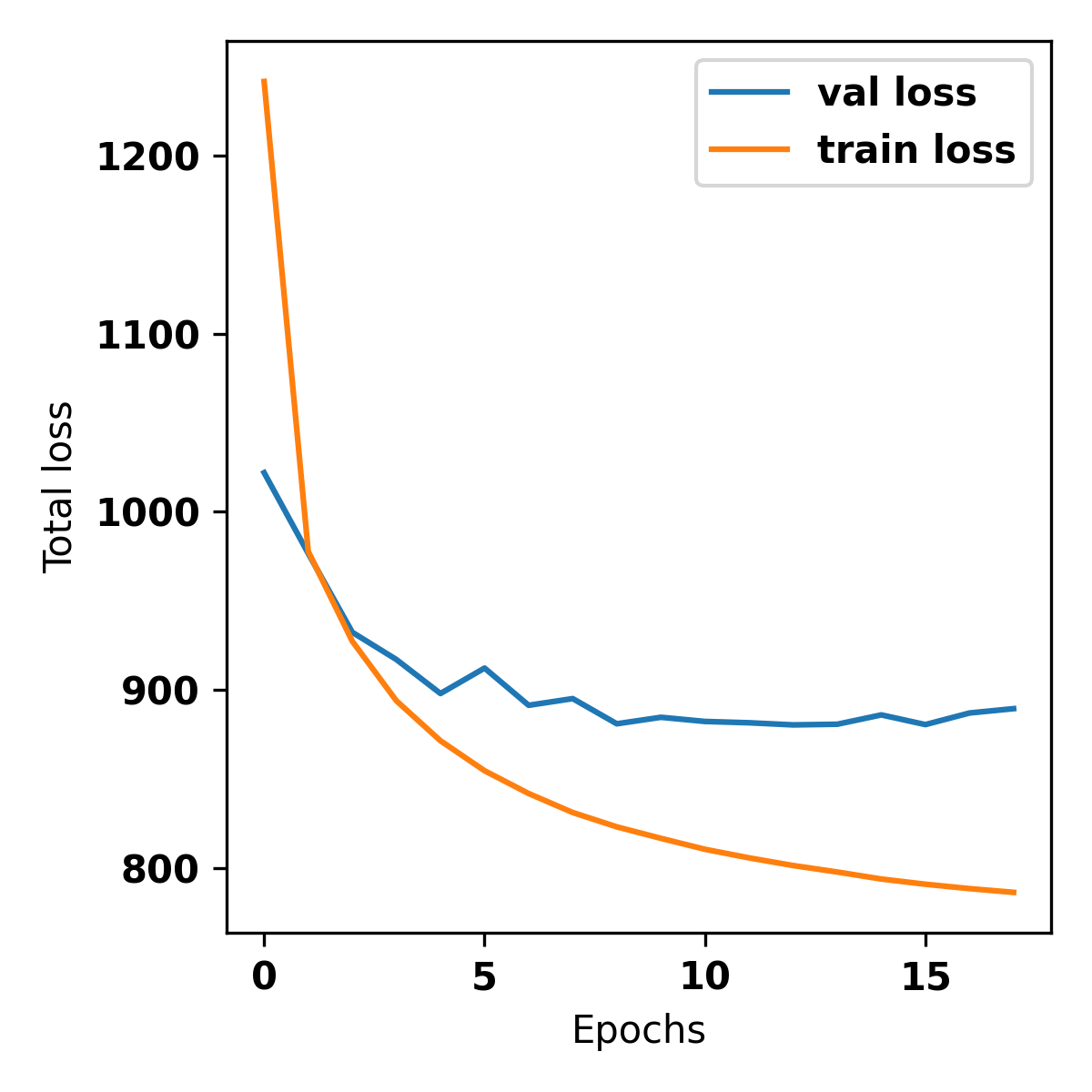
The val loss (validation loss) will decrease and saturate after a few epochs.
Counts Metrics: The pearsonr in peaks should be greater than 0.5 (higher the better). MSE (Mean Squared Error) will be low in peaks.
Profile Metrics: Median JSD (Jensen Shannon Divergence between observed and predicted) lower the better. Median norm JSD is median of the min-max normalized JSD where min JSD is the worst case JSD i.e JSD of observed with uniform profile and max JSD is the best case JSD i.e 0. Median norm JSD is higher the better. Both JSD and median norm JSD are sensitive to read-depth. Higher read-depth results in better metrics.
| peaks.pearsonr | peaks.mse | |
|---|---|---|
| counts_metrics | 0.778526 | 0.646662 |
| peaks.median_jsd | peaks.median_norm_jsd | |
|---|---|---|
| profile_metrics | 0.371181 | 0.429503 |
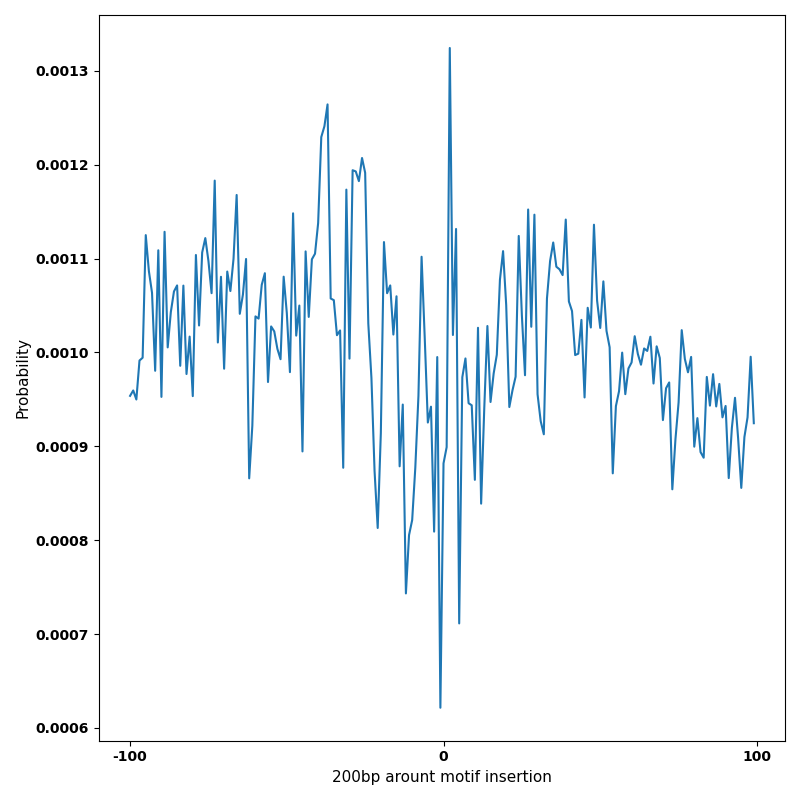
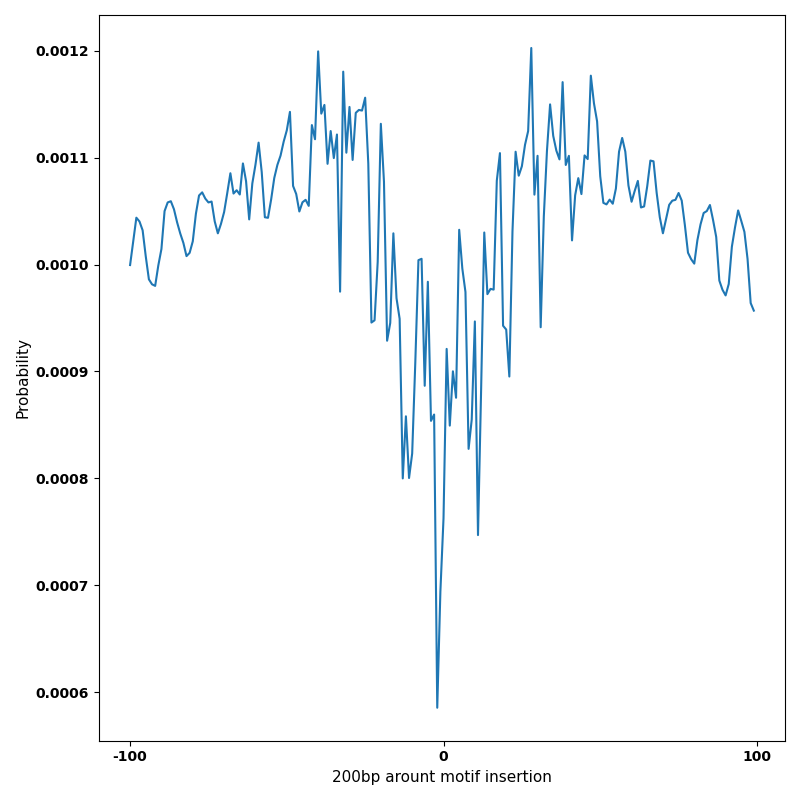
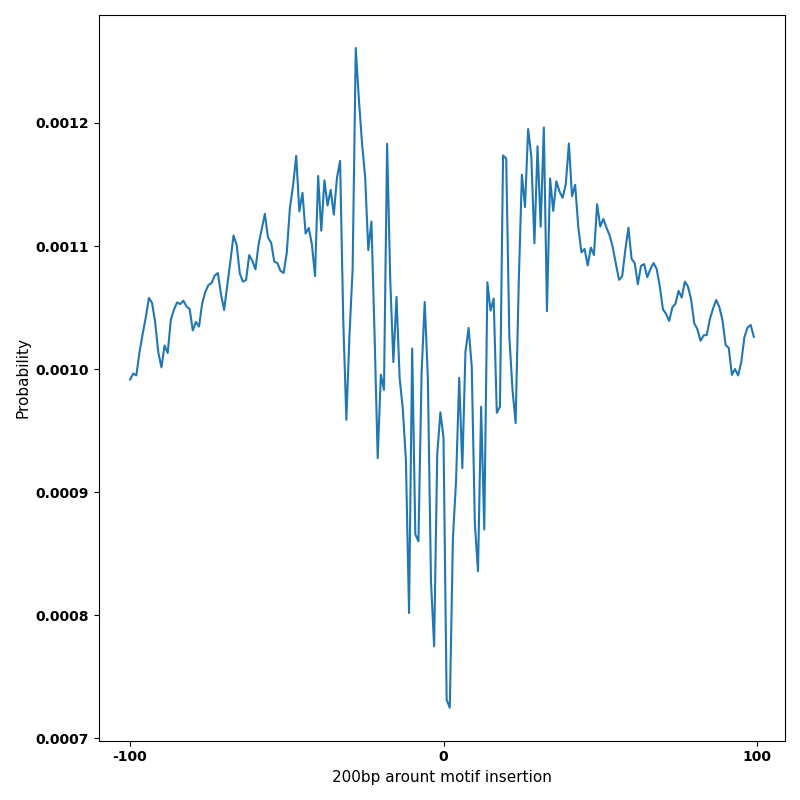
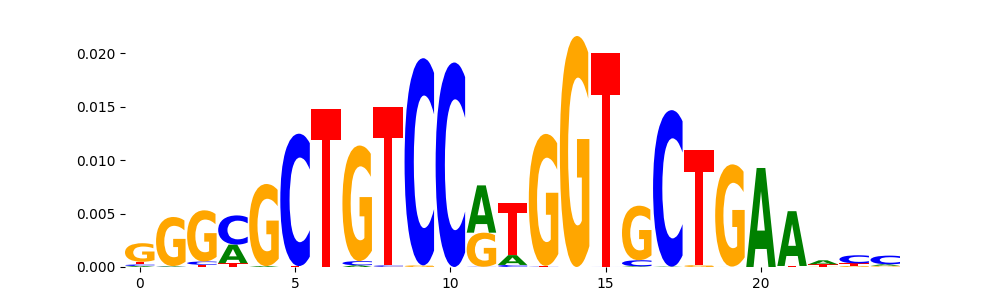
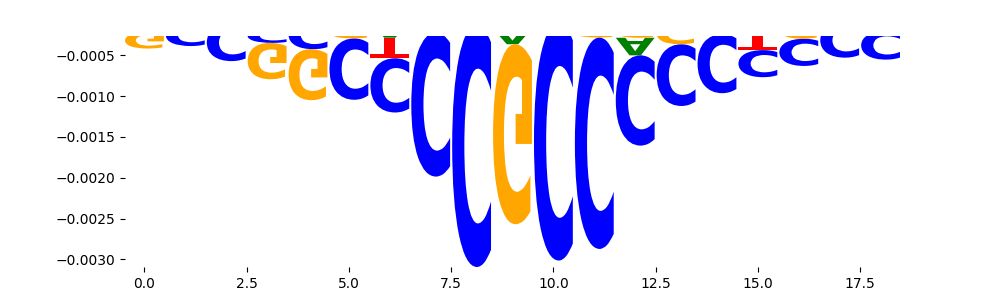
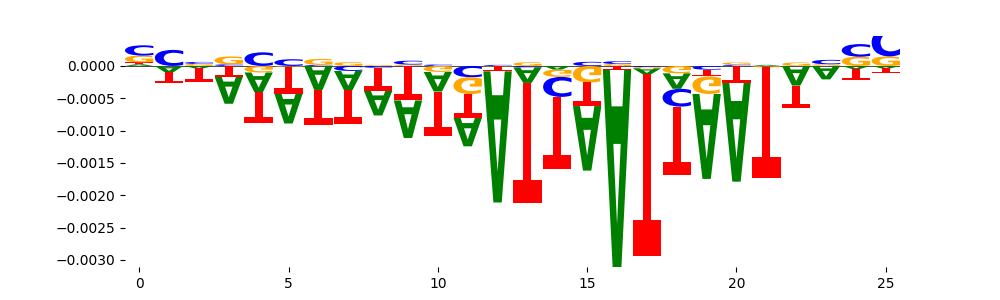
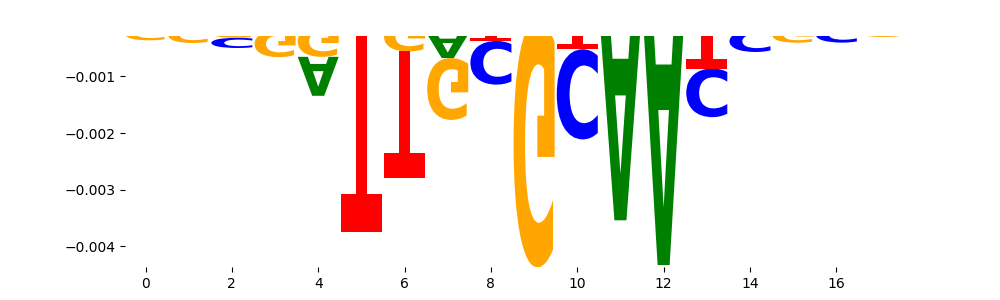
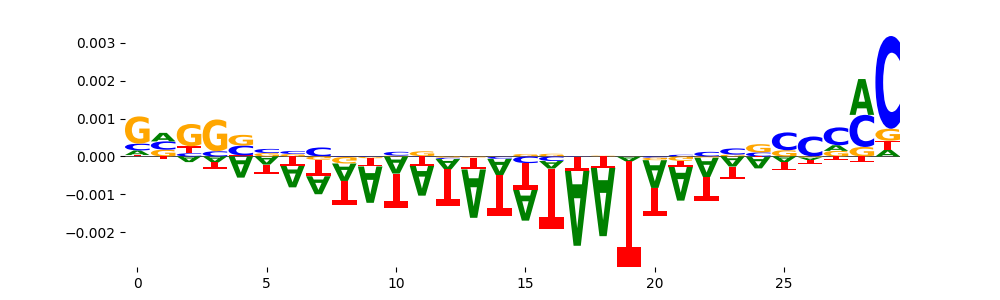
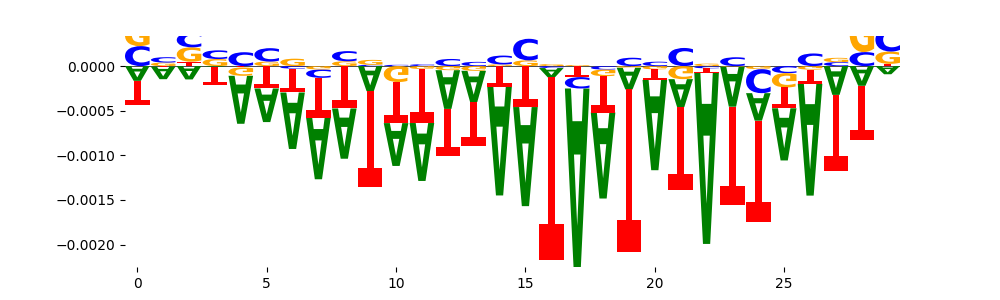
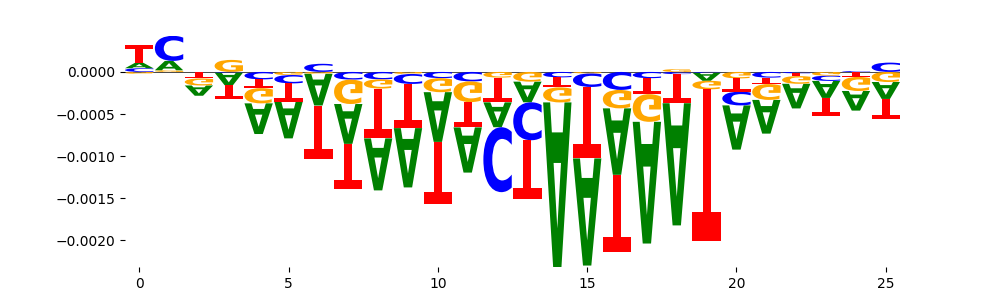
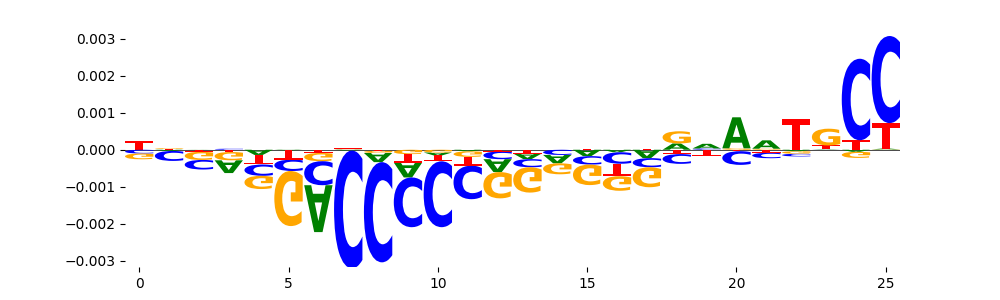
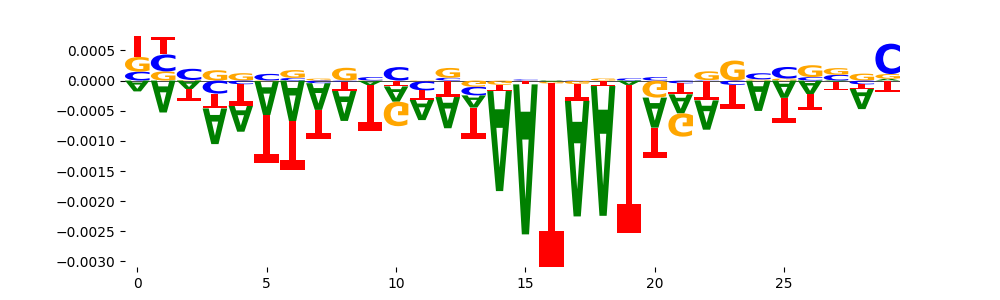
The marginal footprints are the response of the ChromBPNet no bias model to the hetergenous bias motifs. If the bias correction is complete the max of the profiles below should be below 0.003 on all the bias motifs.
For your convenience we calculate here the average of the max of the profiles: 0.001 And the model according to this is corrected
What to do if your model looks uncorrected (i.e max of profiles is greater than 0.003)?
Look at the motifs below captured by TFModisco and you should be able to see motifs that closely look like the bias motifs showing incomplete bias correction. This indicates that your bias model was not completely capturing the response of the bias. We recommend that you use a different pre-trained bias model. For more intuition on choosing the correct pre-trained model or retraining your bias model refer to FAQ section in wiki.
| tn5 motif 1 | tn5 motif 2 | tn5 motif 3 | tn5 motif 4 | tn5 motif 5 |
 |
 |
 |
 |
 |
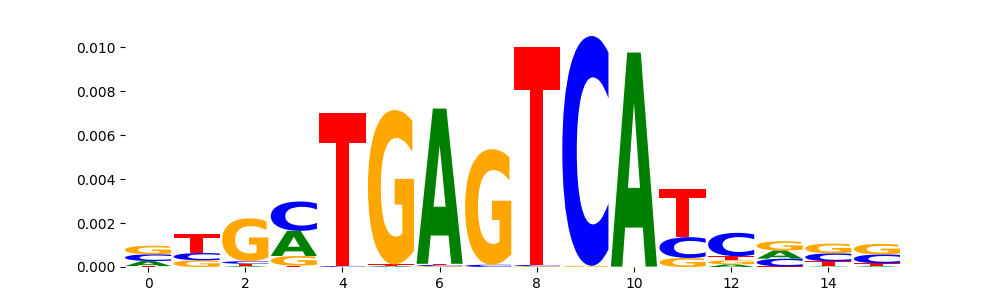
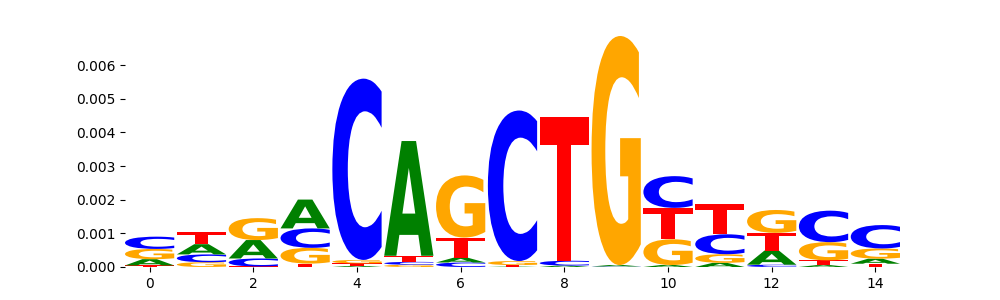
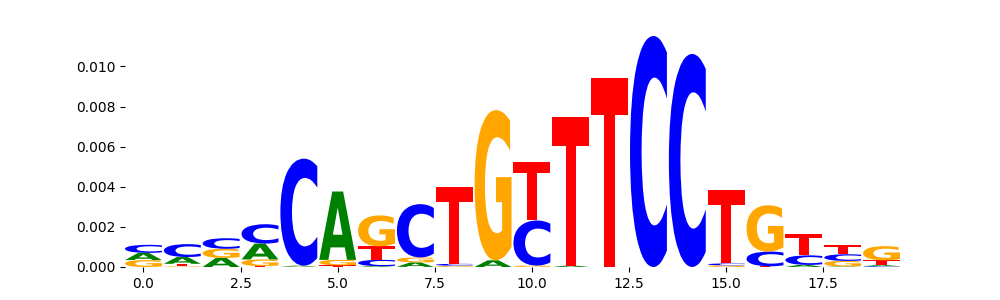
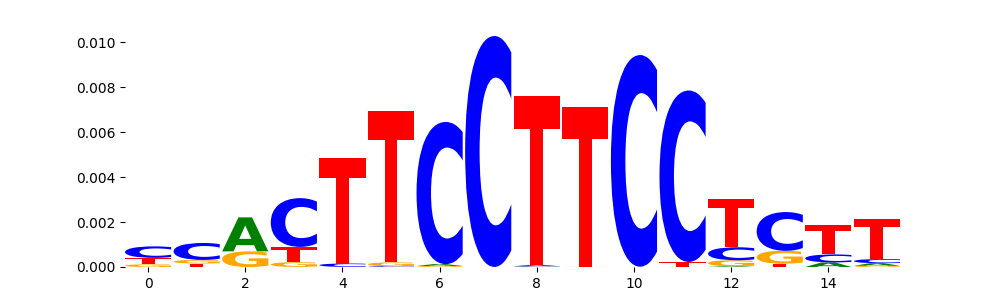
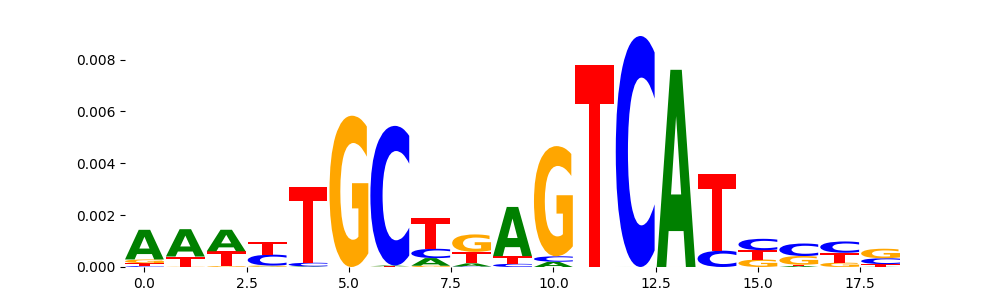
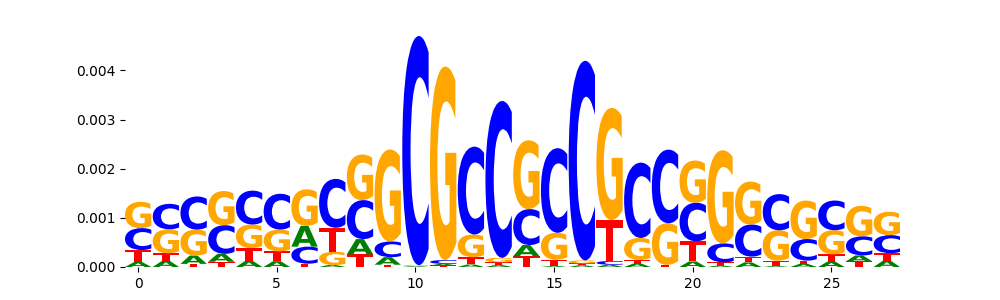
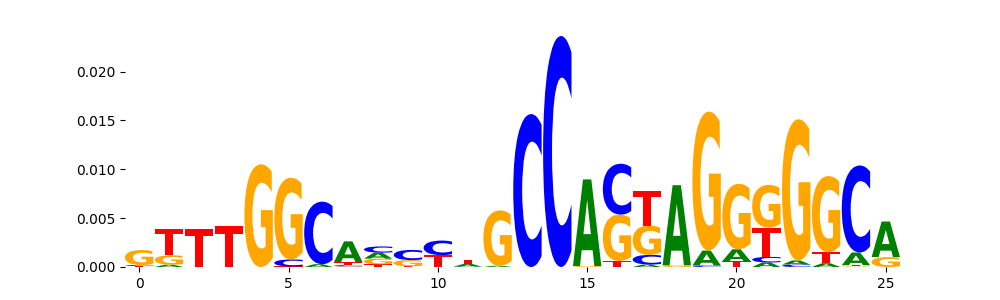
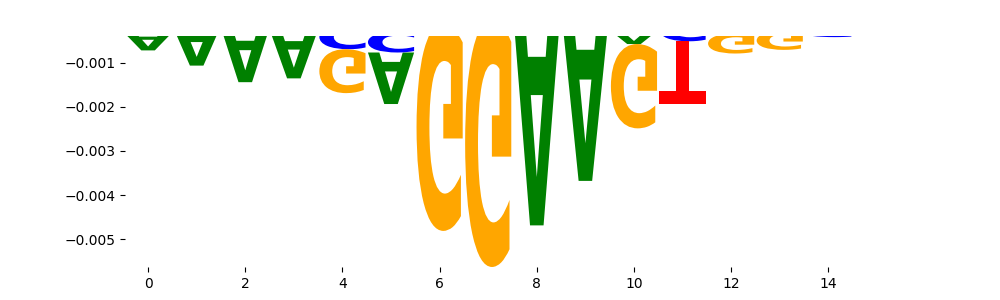
TFModisco motifs generated from profile contribution scores of the ChromBPNet after bias correction model. cwm_fwd, cwm_rev are the forward and reverse complemented consolidated motifs from contribution scores in subset of random peaks. These CWM motifs should be free from any bias motifs and should contain only Transcription Factor (TF) motifs. For each of these motifs, we use TOMTOM to find the top-3 closest matches (match_0, match_1, match_2) from a database consisting of both MEME TF motifs and heterogenous enzyme bias motifs that we have repeatedly seen in our datasets. The qvals (qval0,qval1,qval2) should be low (< 0.0001) for most of the closest TF motif hits (i.e indicating that the closest match is the correct match) - this is also generally verifiable by eye as the closest match will look closely like the CWMs (atleast part of it in case of heterodimers). All the motifs in the list should look nothing like the enzyme bias motif.
What to do if you find an obvious bias motif in the list?
This indicates that your bias model was not completely capturing the response of the bias. We recommend that you use a different pre-trained bias model. For more intuition on choosing the correct pre-trained model or retraining your bias model refer to FAQ section in wiki.
What to do if you find an obvious bias motif in the list?
| pattern | NumSeqs | cwm_fwd | cwm_rev | match0 | qval0 | match0_logo | match1 | qval1 | match1_logo | match2 | qval2 | match2_logo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pos__0 | 6837 | 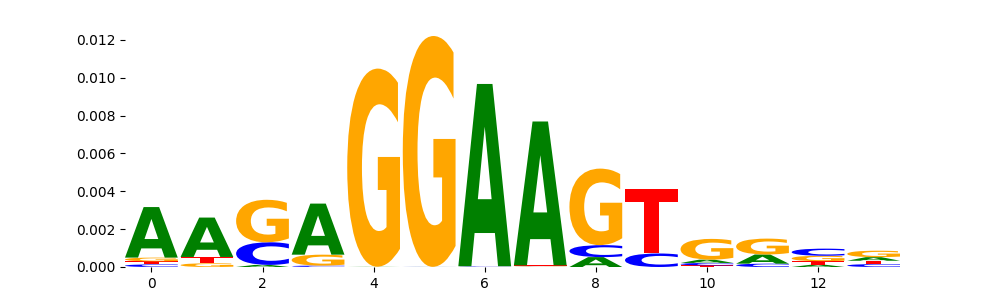 |
 |
ELF5_ETS_1 | 2.216730e-03 |  |
ELF5_ETS_2 | 2.216730e-03 |  |
ELF5_MA0136.2 | 2.216730e-03 |  |
| pos__1 | 4224 |  |
 |
CTCF_MA0139.1 | 1.178970e-13 | 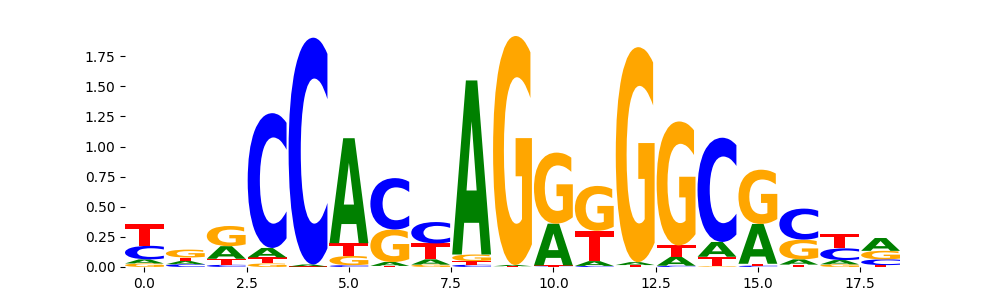 |
CTCF_HUMAN.H11MO.0.A | 2.056560e-10 | 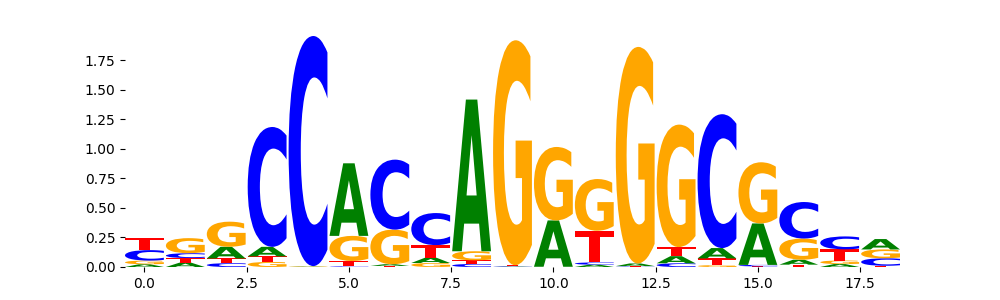 |
CTCF_MOUSE.H11MO.0.A | 1.576290e-09 | 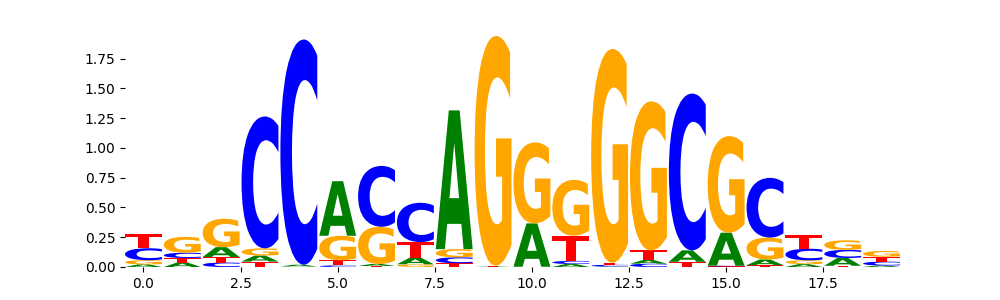 |
| pos__2 | 2903 | 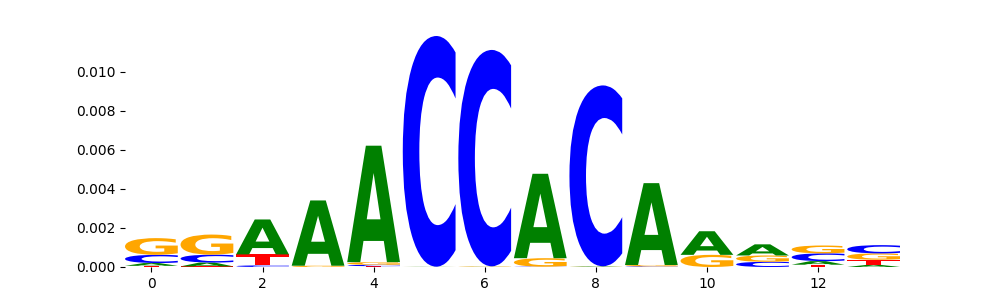 |
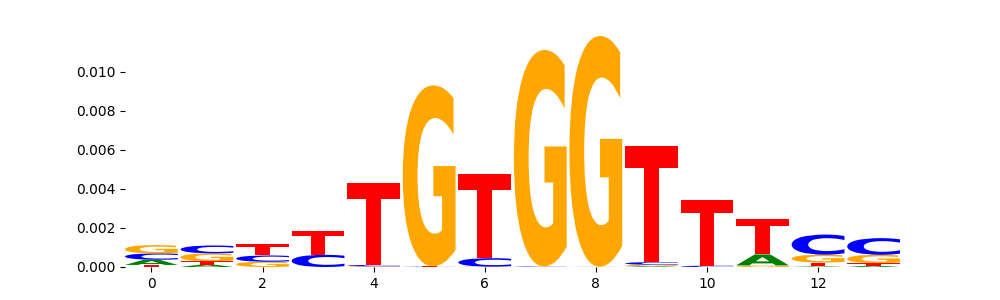 |
RUNX1_HUMAN.H11MO.0.A | 4.091430e-03 | 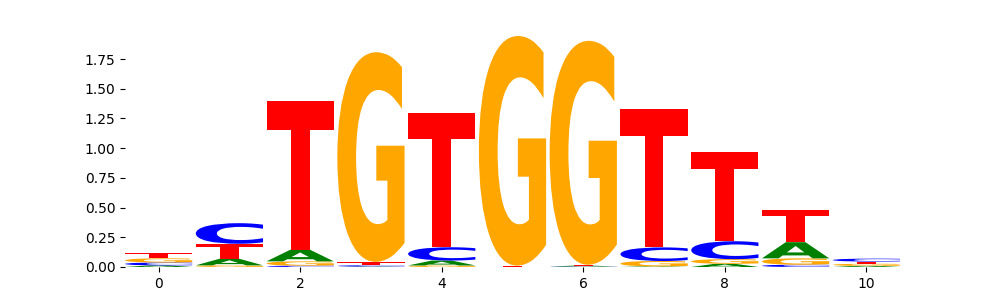 |
RUNX1_MOUSE.H11MO.0.A | 4.091430e-03 |  |
RUNX3_HUMAN.H11MO.0.A | 5.978970e-02 | 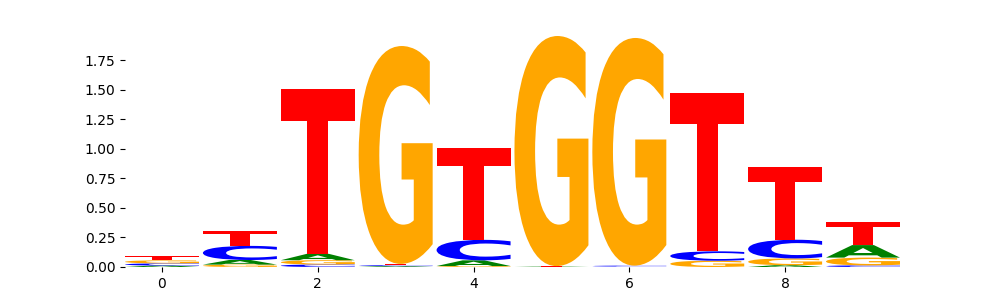 |
| pos__3 | 1736 |  |
 |
CEBPA_MA0102.3 | 2.567930e-09 |  |
CEBPB_HUMAN.H11MO.0.A | 8.610760e-09 |  |
CEBPB_MOUSE.H11MO.0.A | 4.721960e-08 |  |
| pos__4 | 1435 |  |
 |
JUN_HUMAN.H11MO.0.A | 2.718640e-05 |  |
FOSL2_HUMAN.H11MO.0.A | 2.718640e-05 | 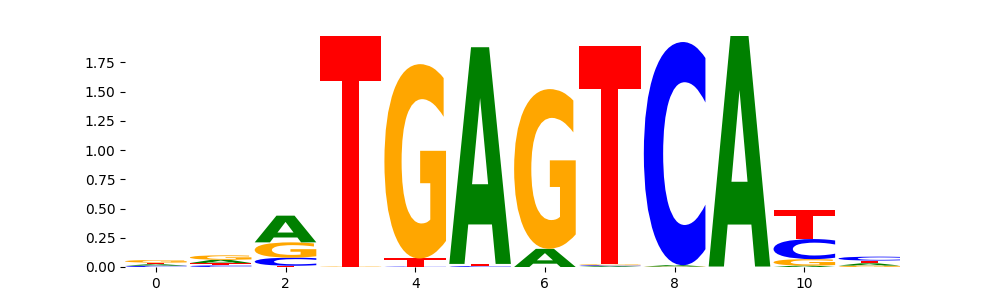 |
FOS_MOUSE.H11MO.0.A | 1.074530e-04 |  |
| pos__5 | 1159 | 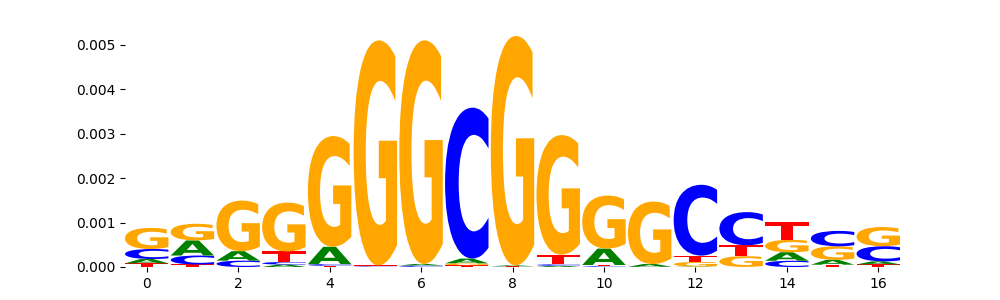 |
 |
KLF12_HUMAN.H11MO.0.C | 5.334890e-05 |  |
SP3_HUMAN.H11MO.0.B | 7.113190e-05 | 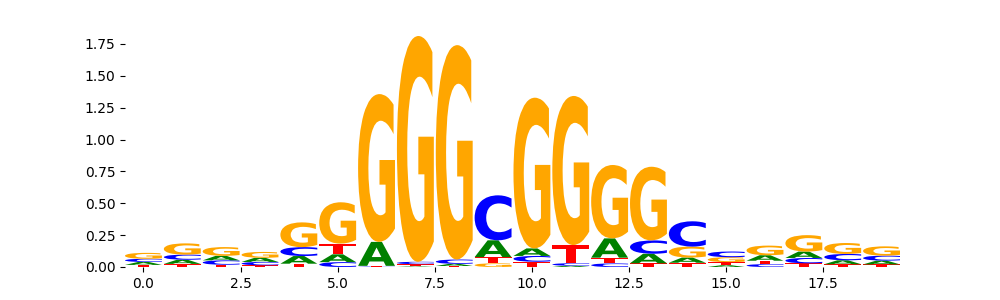 |
SP3_MOUSE.H11MO.0.B | 7.113190e-05 | 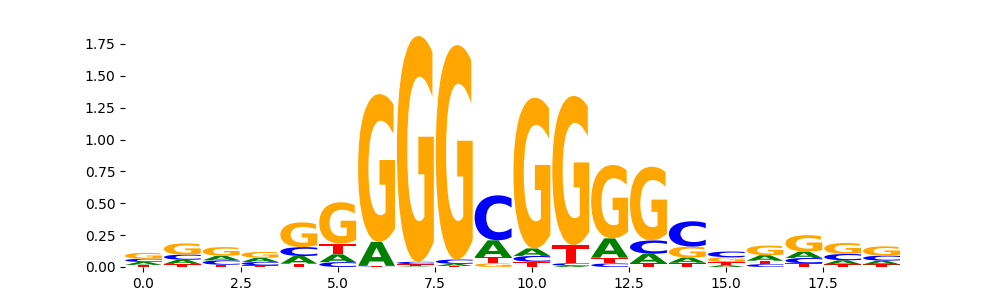 |
| pos__6 | 1058 |  |
 |
IRF1_MOUSE.H11MO.0.A | 1.963380e-07 | 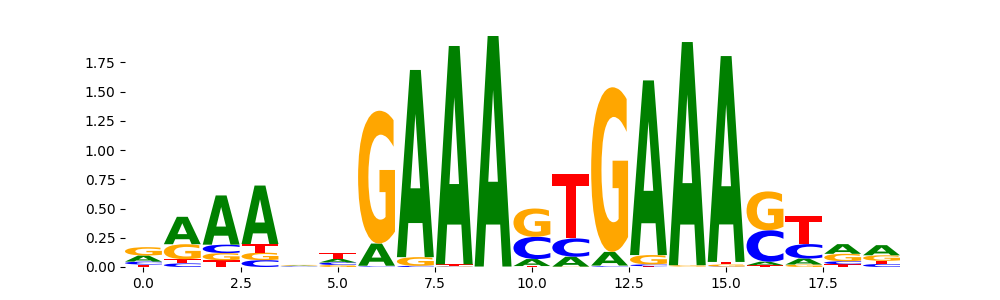 |
IRF1_MA0050.2 | 1.445590e-06 |  |
IRF1_HUMAN.H11MO.0.A | 1.708240e-06 | 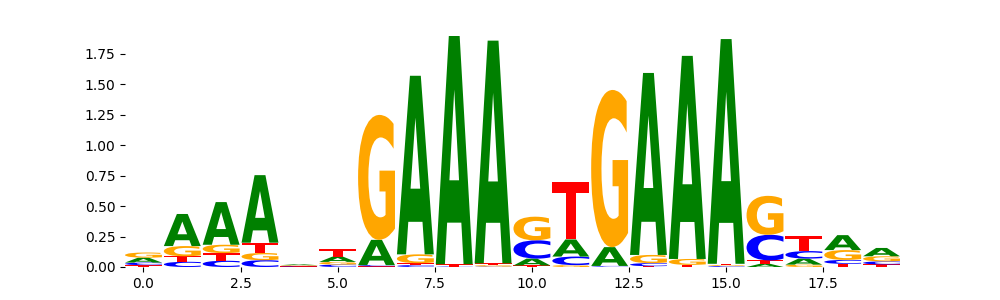 |
| pos__7 | 1009 | 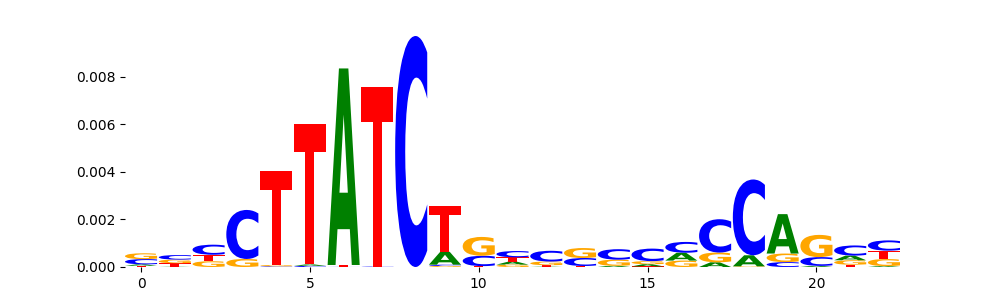 |
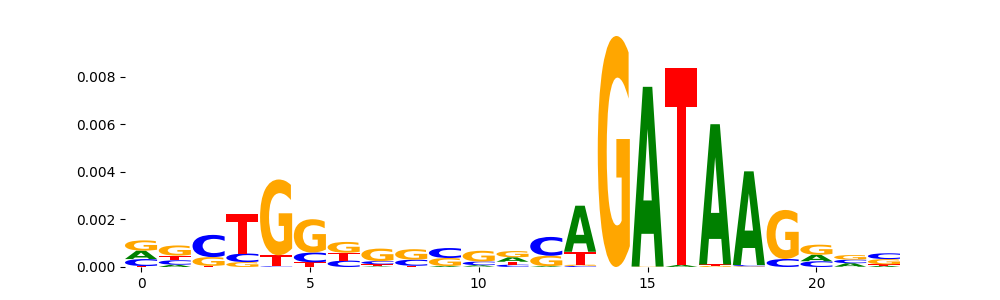 |
GATA2_HUMAN.H11MO.0.A | 4.291870e-10 | 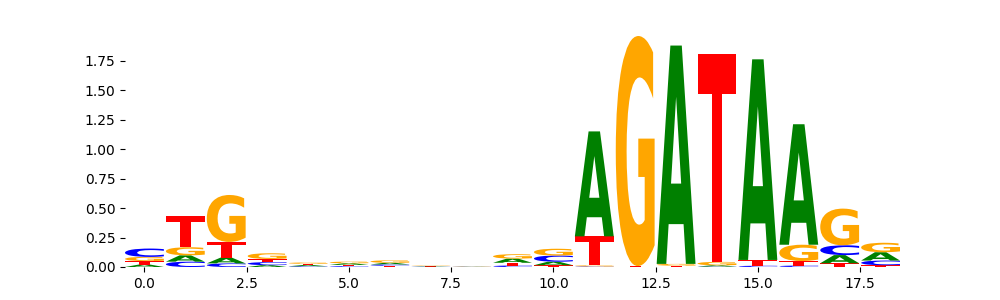 |
TAL1_MOUSE.H11MO.0.A | 1.958300e-08 | 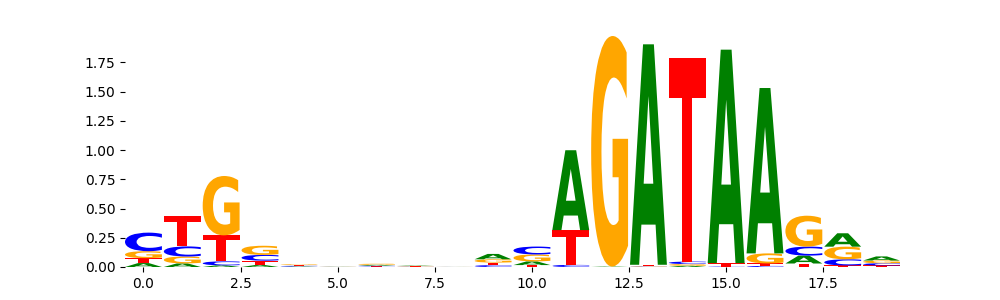 |
GATA1_HUMAN.H11MO.0.A | 1.643580e-04 | 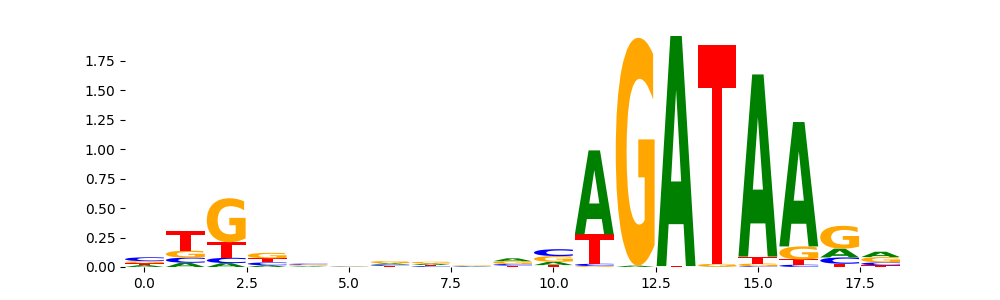 |
| pos__8 | 815 |  |
 |
ELK1_ETS_1 | 9.109300e-02 | 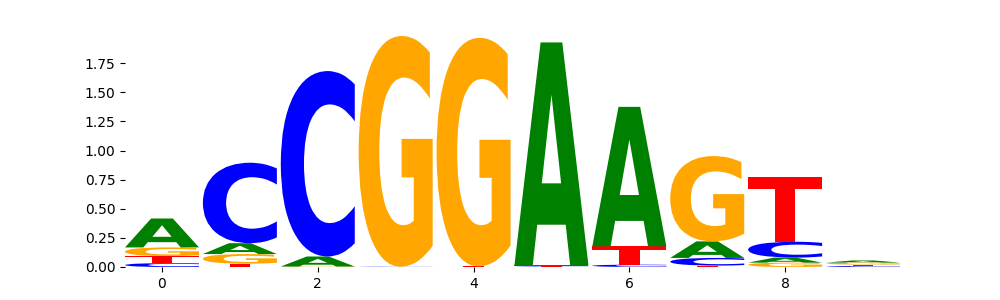 |
ELK1_ETS_2 | 9.109300e-02 | 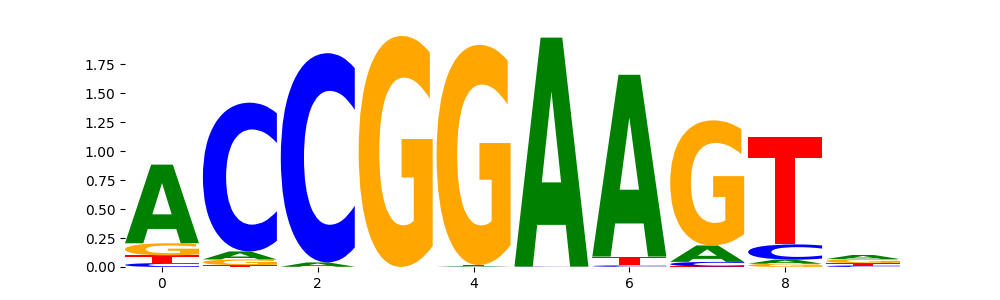 |
ELK1_ETS_4 | 9.109300e-02 | 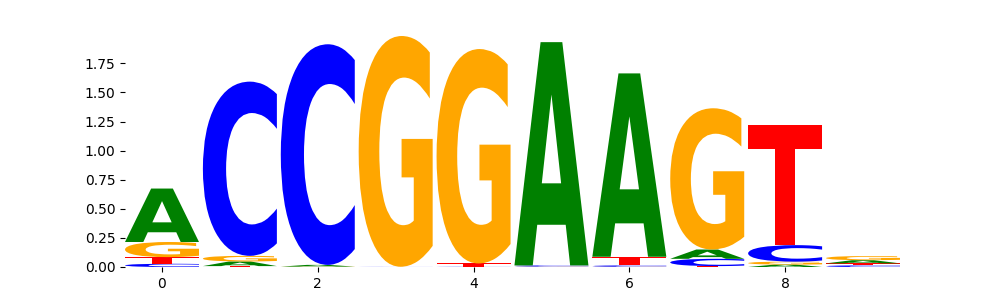 |
| pos__9 | 740 | 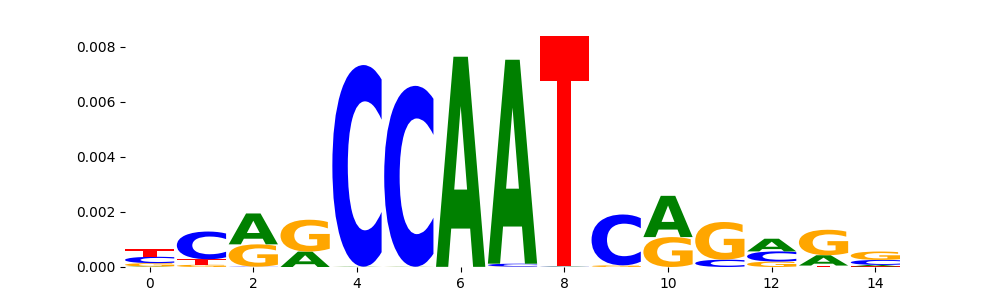 |
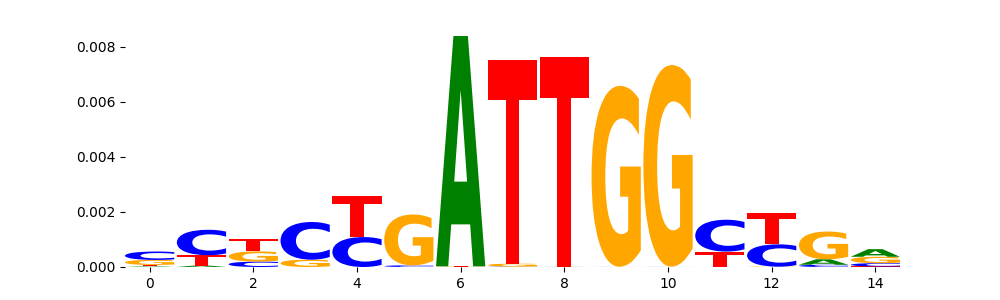 |
NFYB_HUMAN.H11MO.0.A | 5.050080e-04 | 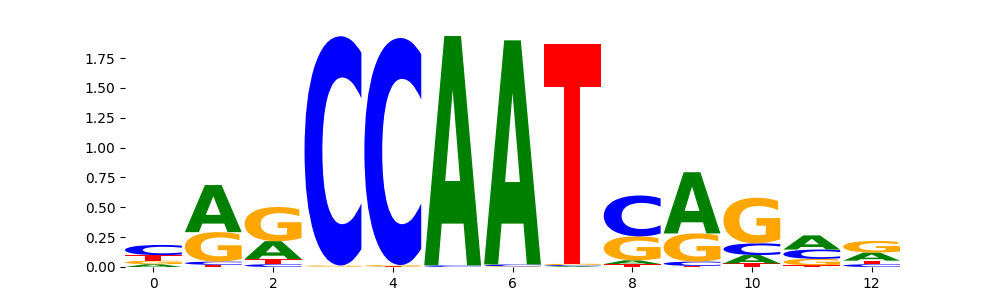 |
NFYB_MOUSE.H11MO.0.A | 5.050080e-04 |  |
NFYC_HUMAN.H11MO.0.A | 5.050080e-04 | 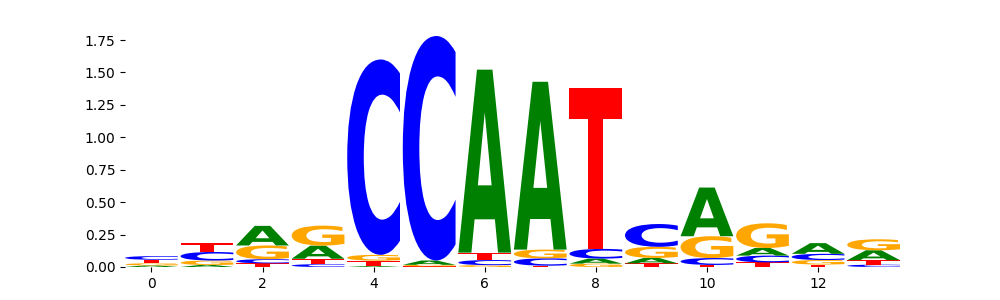 |
| pos__10 | 735 | 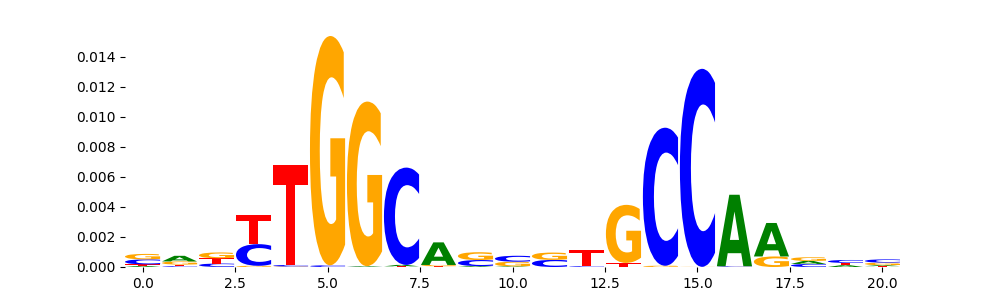 |
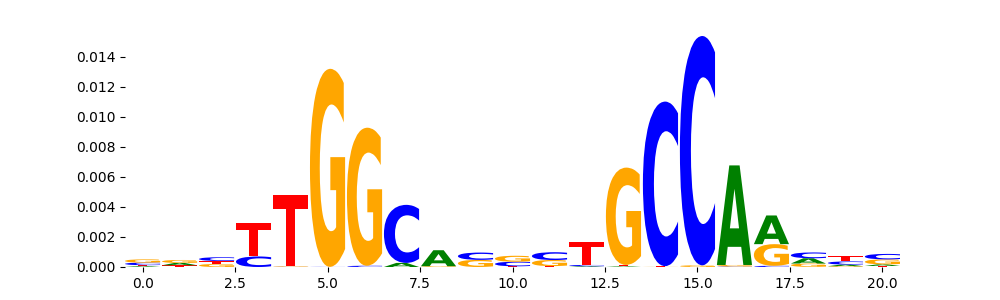 |
NFIA_HUMAN.H11MO.0.C | 4.607910e-07 | 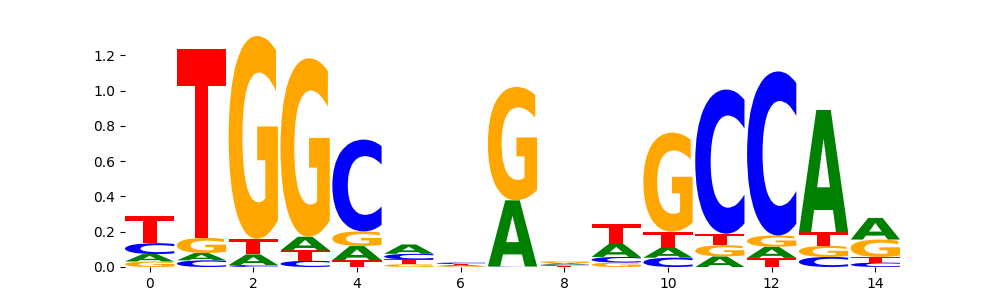 |
NFIA_MOUSE.H11MO.0.C | 4.607910e-07 |  |
NFIC_HUMAN.H11MO.0.A | 2.353280e-05 |  |
| pos__11 | 725 | 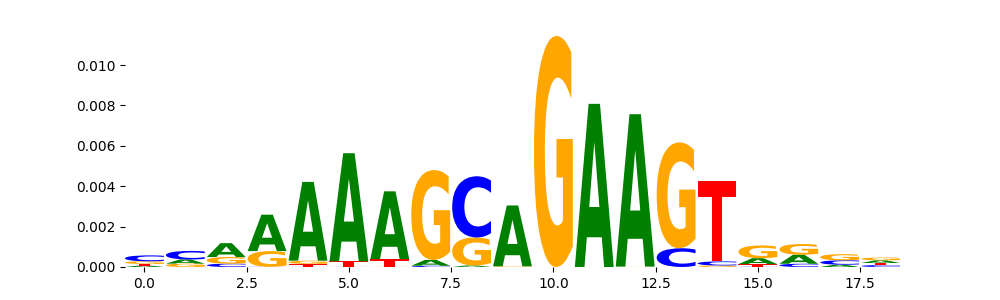 |
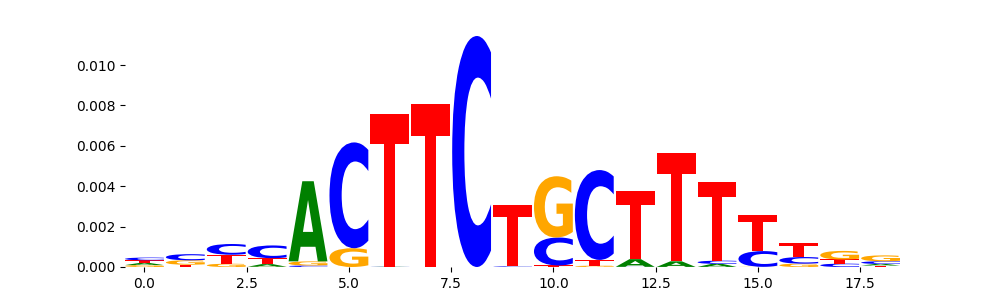 |
SPI1_ETS_1 | 1.965340e-02 | 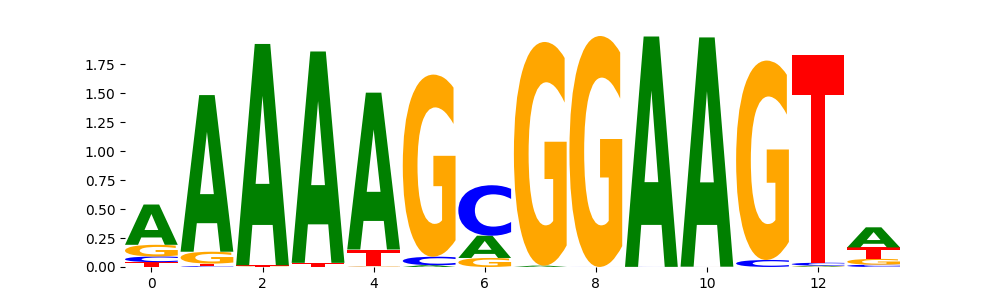 |
SPI1_MA0080.4 | 1.965340e-02 | 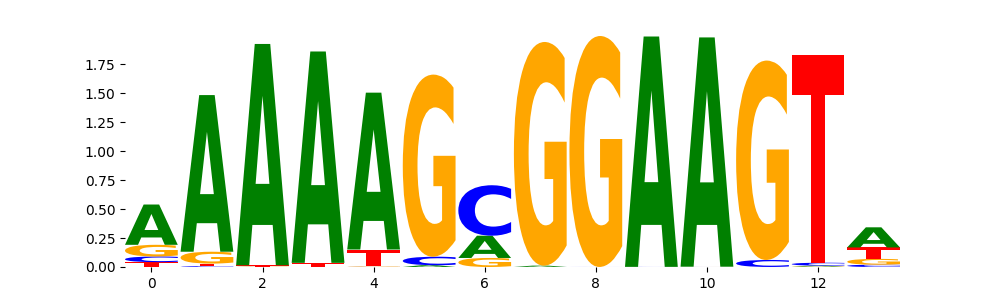 |
SPIB_ETS_1 | 1.965340e-02 | 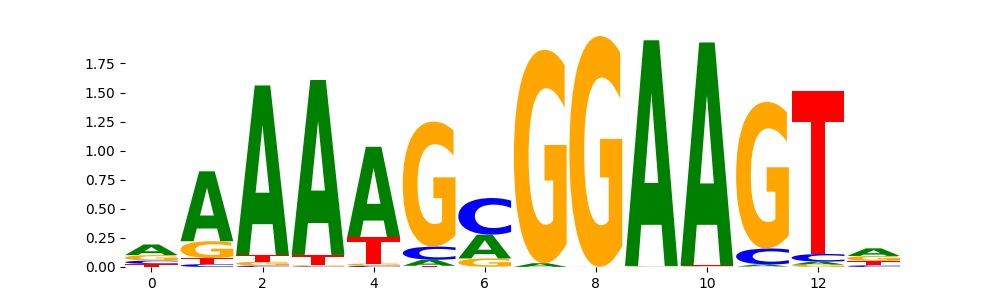 |
| pos__12 | 602 |  |
 |
ATF1_HUMAN.H11MO.0.B | 3.696830e-04 |  |
CREB1_HUMAN.H11MO.0.A | 3.696830e-04 |  |
CREB1_MOUSE.H11MO.0.A | 3.696830e-04 |  |
| pos__13 | 507 |  |
 |
OLIG2_HUMAN.H11MO.0.B | 1.278540e-03 | 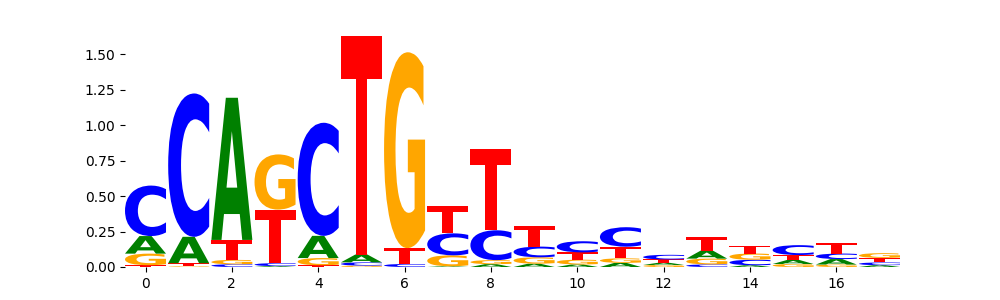 |
OLIG2_MOUSE.H11MO.0.A | 1.278540e-03 | 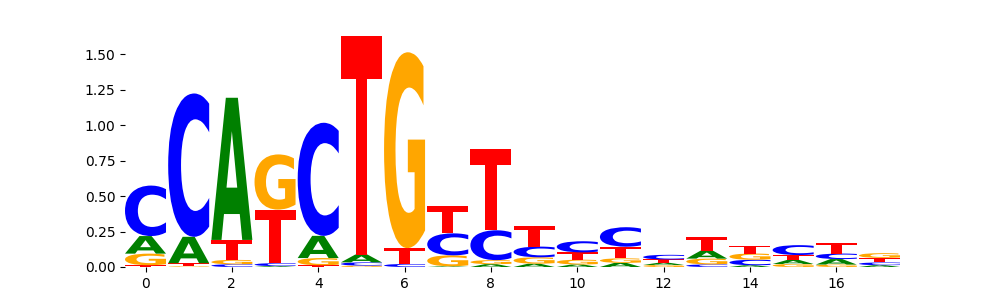 |
LYL1_HUMAN.H11MO.0.A | 1.570500e-03 |  |
| pos__14 | 466 |  |
 |
LYL1_HUMAN.H11MO.0.A | 2.350970e-05 |  |
LYL1_MOUSE.H11MO.0.A | 2.350970e-05 | 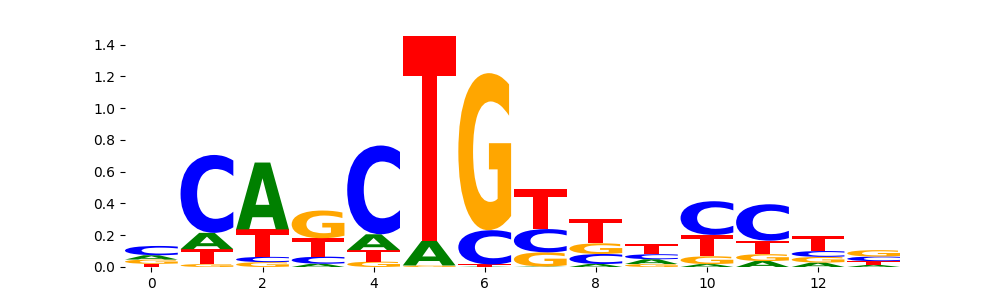 |
OLIG2_HUMAN.H11MO.0.B | 1.096260e-03 |  |
| pos__15 | 451 | 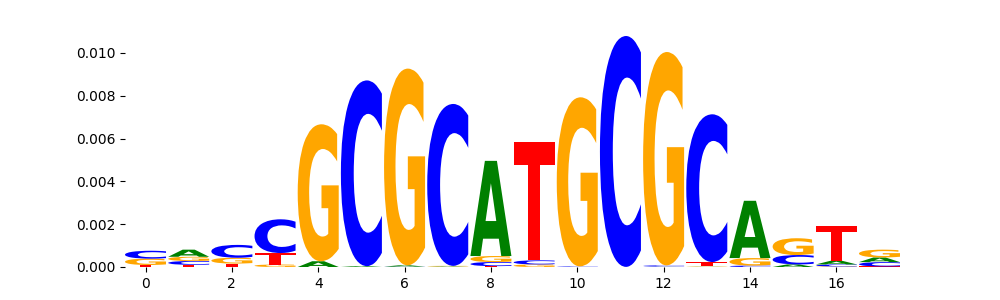 |
 |
NRF1_MOUSE.H11MO.0.A | 2.216940e-08 | 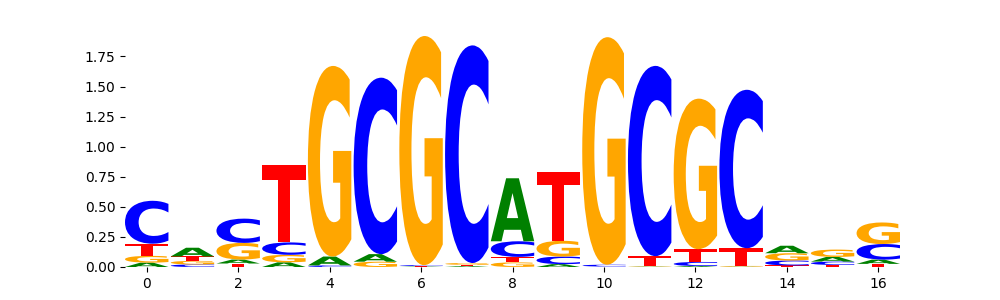 |
NRF1_HUMAN.H11MO.0.A | 7.299730e-07 |  |
NRF1_NRF_1 | 2.131500e-05 |  |
| pos__16 | 388 |  |
 |
EWSR1-FLI1_MA0149.1 | 6.639900e-03 | 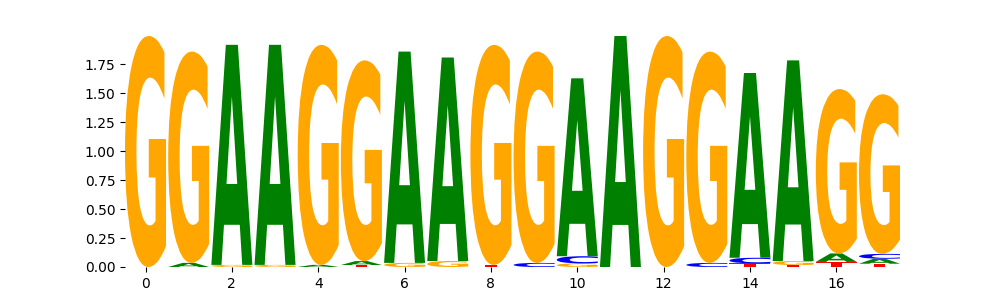 |
FLI1_HUMAN.H11MO.0.A | 3.633030e-01 | 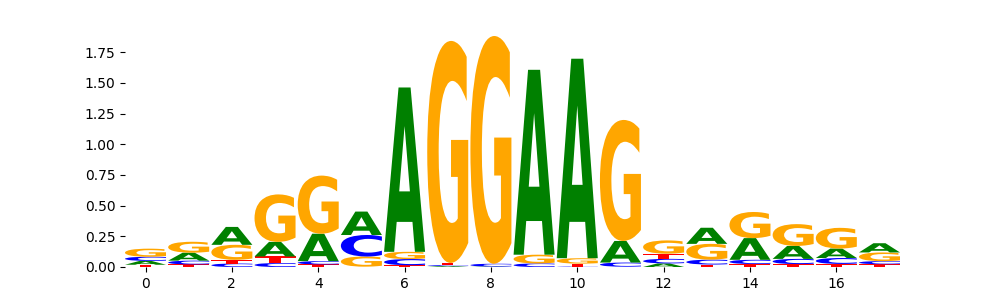 |
ETS2_HUMAN.H11MO.0.B | 4.373020e-01 | 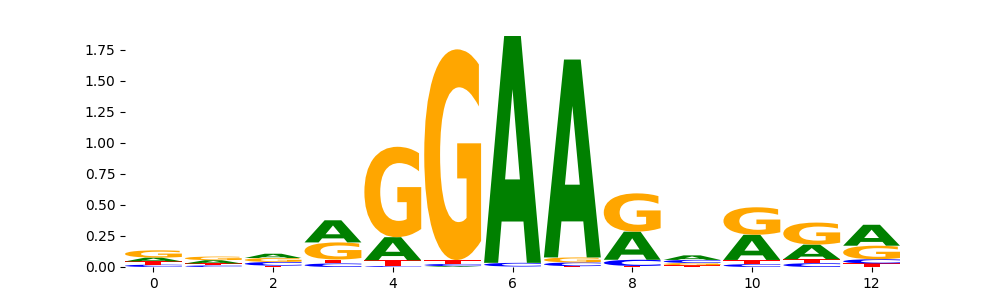 |
| pos__17 | 361 |  |
 |
BACH2_HUMAN.H11MO.0.A | 2.393550e-03 |  |
BACH2_MOUSE.H11MO.0.A | 2.393550e-03 | 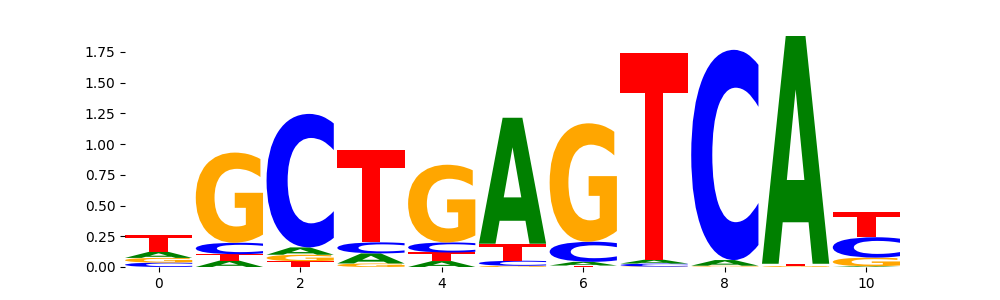 |
NF2L2_MOUSE.H11MO.0.A | 2.393550e-03 |  |
| pos__18 | 344 | 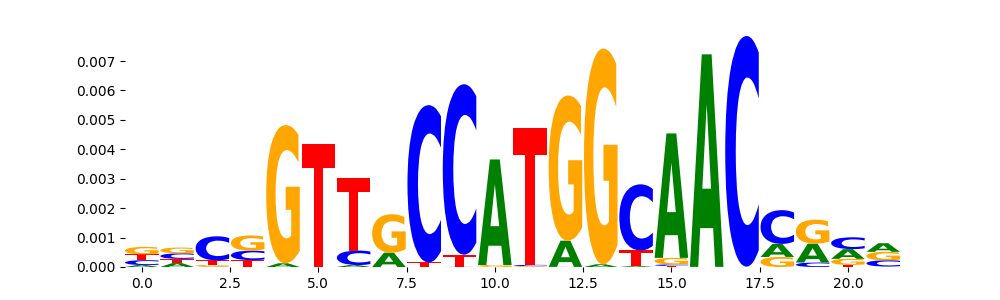 |
 |
RFX3_MA0798.1 | 2.523330e-09 |  |
RFX3_RFX_1 | 2.523330e-09 | 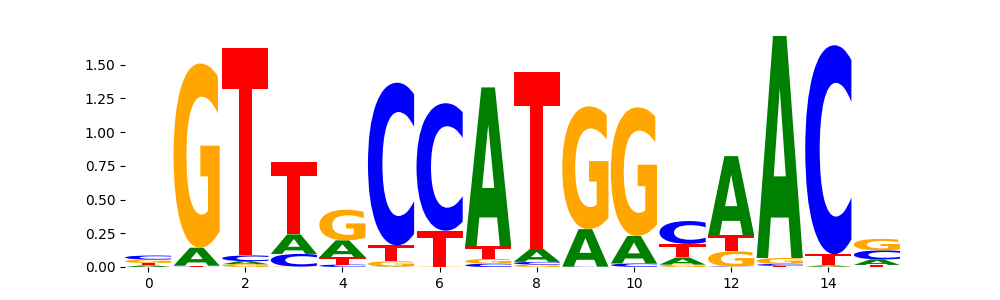 |
RFX2_MA0600.2 | 3.307830e-09 |  |
| pos__19 | 343 |  |
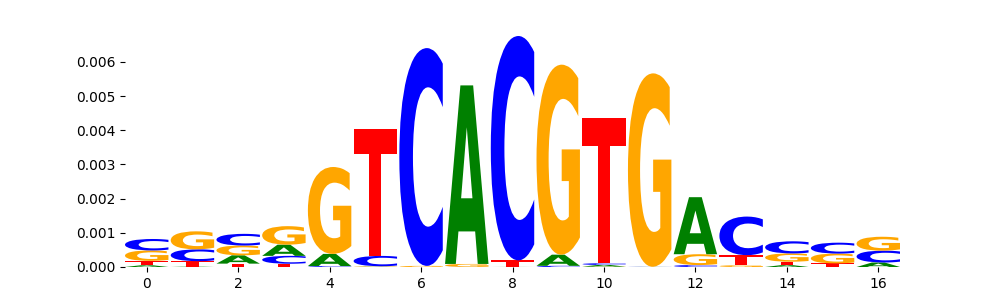 |
MITF_HUMAN.H11MO.0.A | 1.392410e-05 |  |
USF1_HUMAN.H11MO.0.A | 3.720380e-05 |  |
TFE3_bHLH_1 | 3.720380e-05 |  |
| pos__20 | 285 | 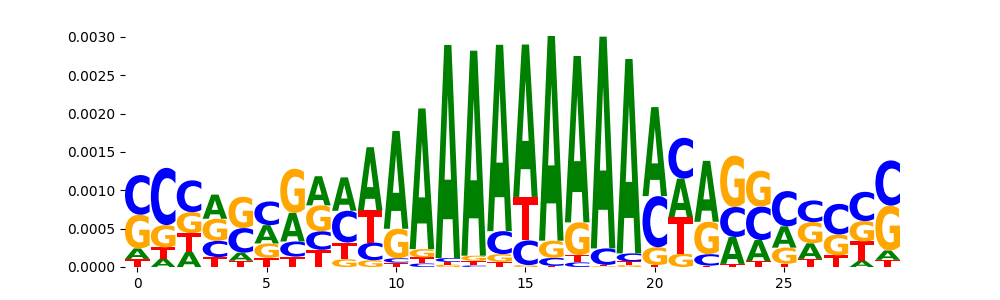 |
 |
ZNF384_MA1125.1 | 1.621800e-02 |  |
PRDM6_HUMAN.H11MO.0.C | 3.185130e-02 |  |
ANDR_HUMAN.H11MO.0.A | 8.486880e-02 |  |
| pos__21 | 264 |  |
 |
SP2_HUMAN.H11MO.0.A | 4.128050e-02 | 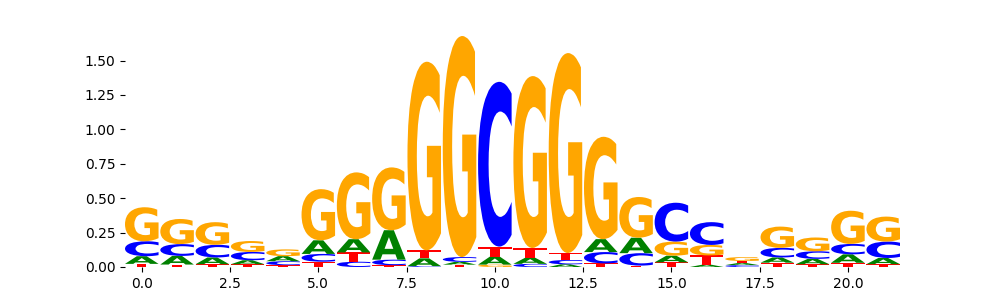 |
SP2_MOUSE.H11MO.0.B | 4.128050e-02 | 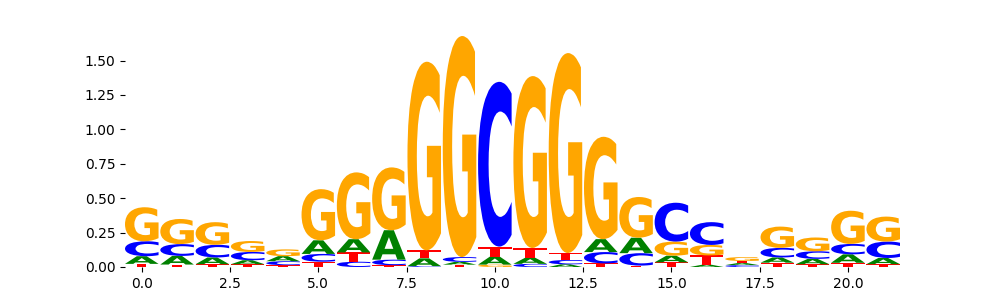 |
SP3_HUMAN.H11MO.0.B | 4.574220e-02 |  |
| pos__22 | 255 |  |
 |
SP2_HUMAN.H11MO.0.A | 5.509340e-06 |  |
SP2_MOUSE.H11MO.0.B | 5.509340e-06 |  |
SP1_MOUSE.H11MO.0.A | 1.063590e-05 | 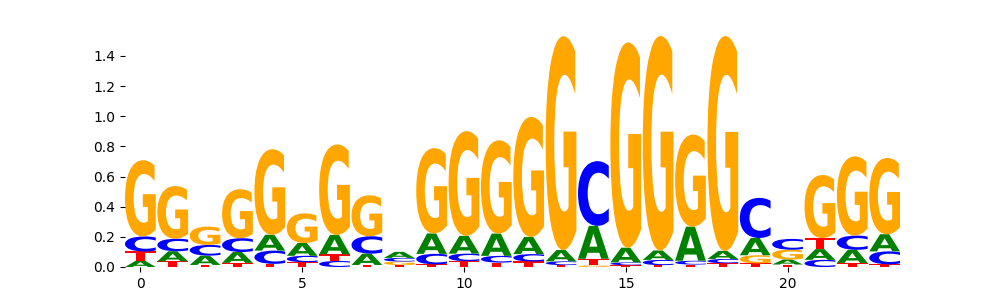 |
| pos__23 | 186 | 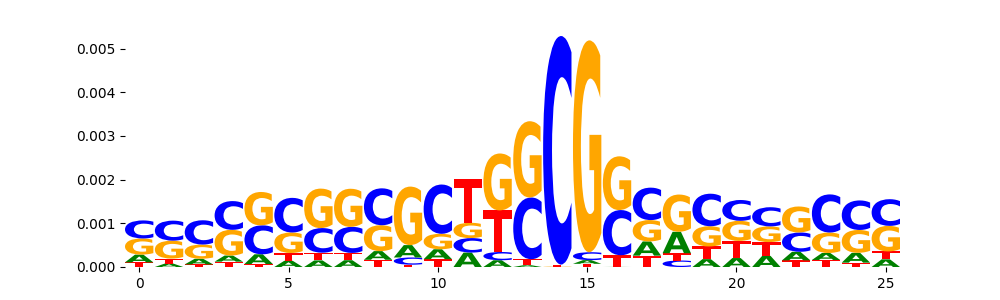 |
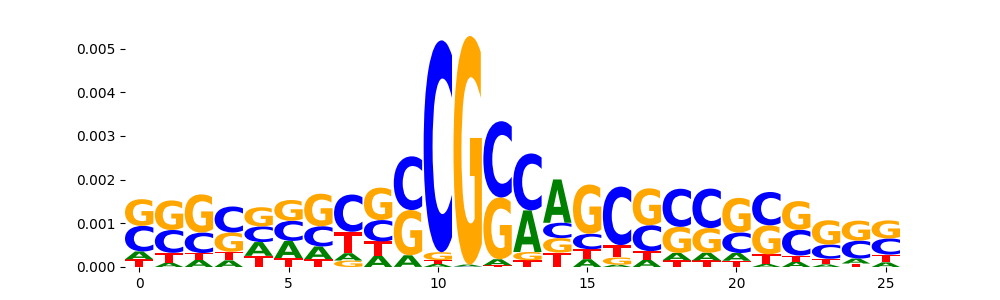 |
SP2_HUMAN.H11MO.0.A | 6.402330e-03 |  |
SP2_MOUSE.H11MO.0.B | 6.402330e-03 |  |
ZFX_MOUSE.H11MO.0.B | 1.608600e-02 | 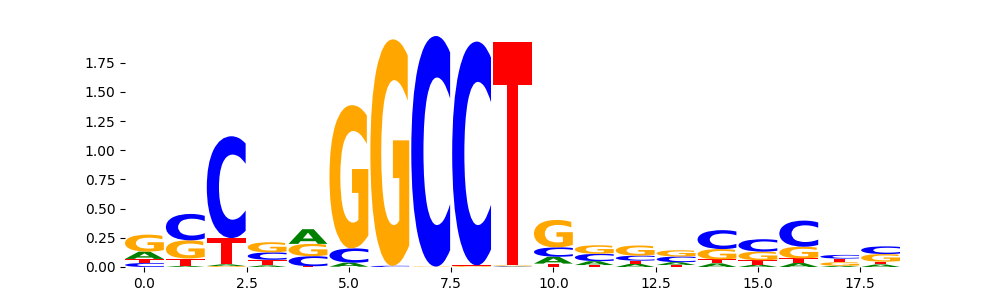 |
| pos__24 | 175 | 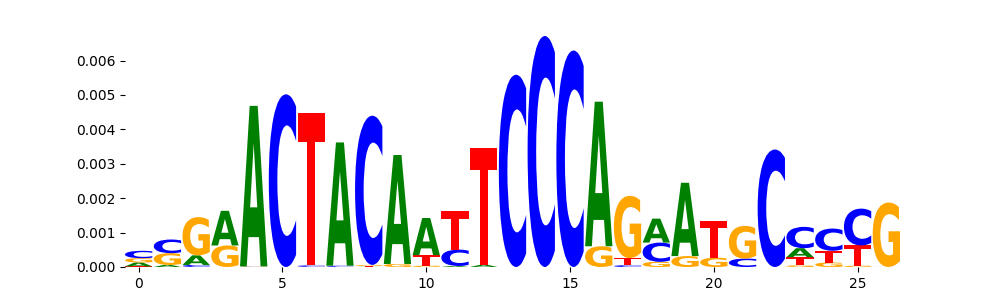 |
 |
ZNF76_HUMAN.H11MO.0.C | 8.617420e-22 | 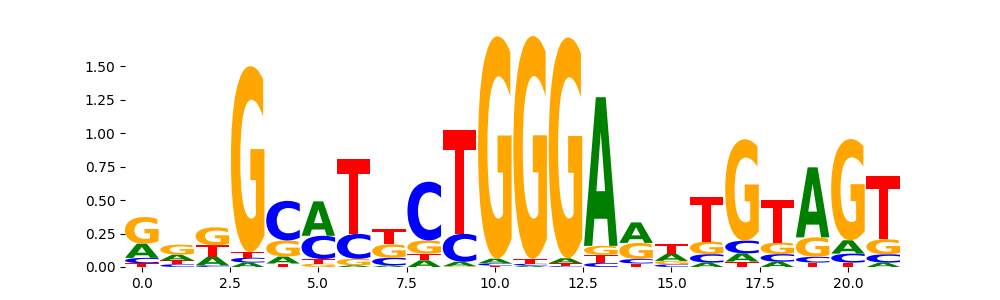 |
ZN143_HUMAN.H11MO.0.A | 2.092080e-19 | 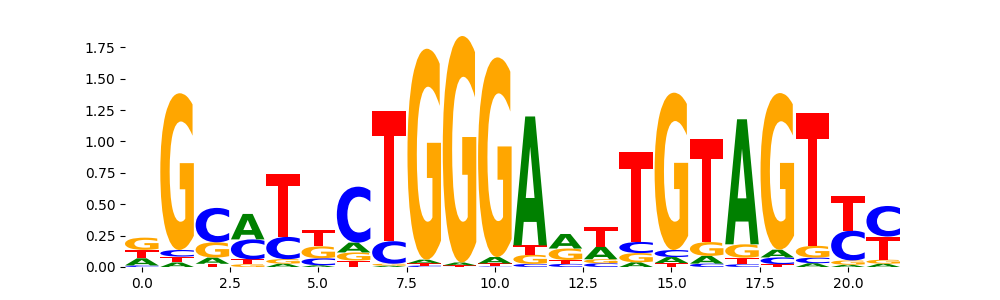 |
THA11_MOUSE.H11MO.0.B | 7.550920e-19 | 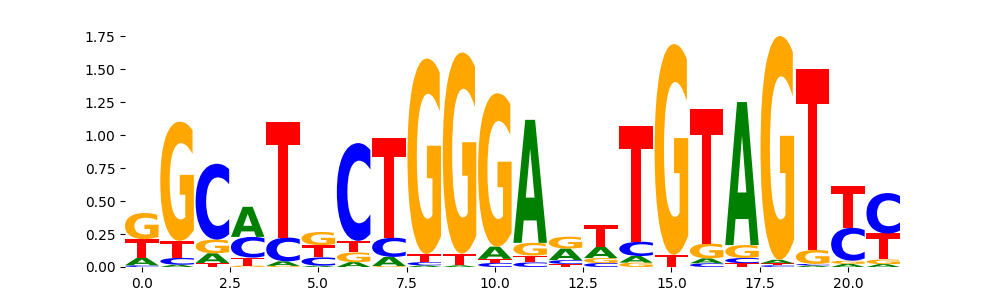 |
| pos__25 | 160 | 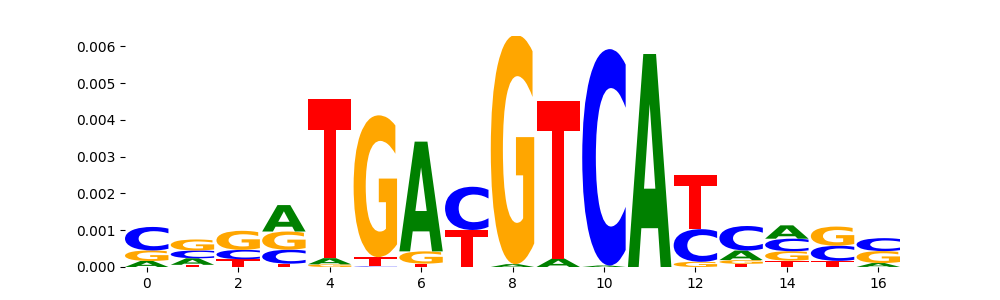 |
 |
JUN_MA0488.1 | 1.007840e-02 |  |
ATF2_HUMAN.H11MO.0.B | 1.007840e-02 |  |
ATF2_MOUSE.H11MO.0.A | 1.007840e-02 |  |
| pos__26 | 150 | 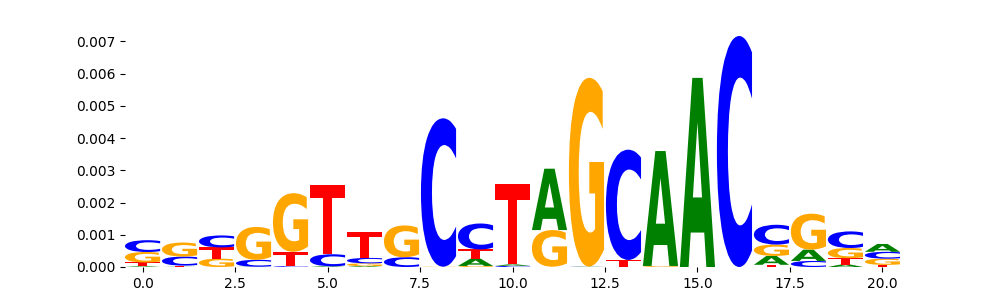 |
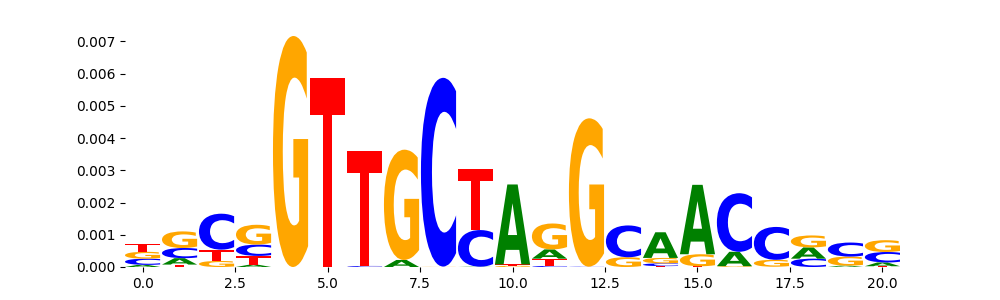 |
RFX2_RFX_2 | 8.812530e-07 |  |
RFX4_RFX_2 | 8.812530e-07 | 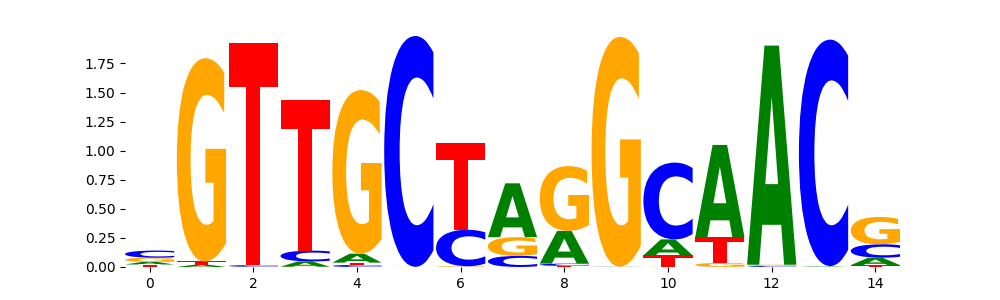 |
Rfx2.mouse_RFX_2 | 8.812530e-07 | 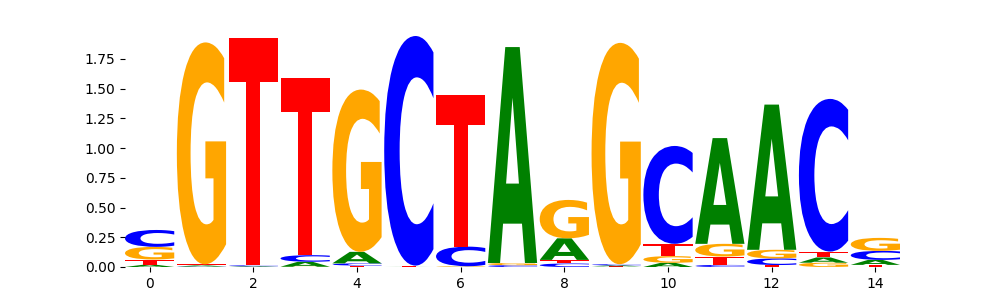 |
| pos__27 | 133 | 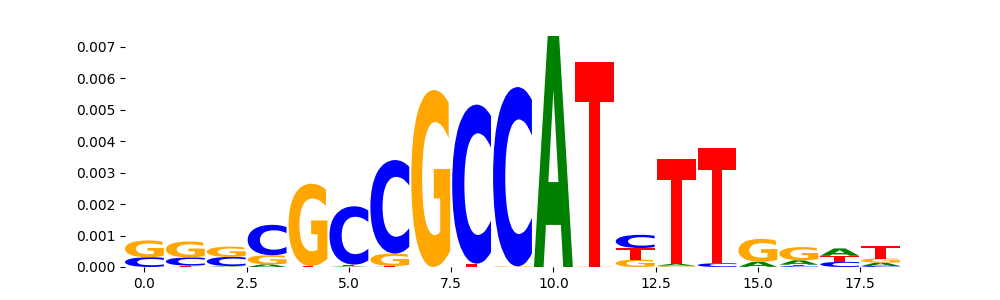 |
 |
TYY1_MOUSE.H11MO.0.A | 6.776570e-06 | 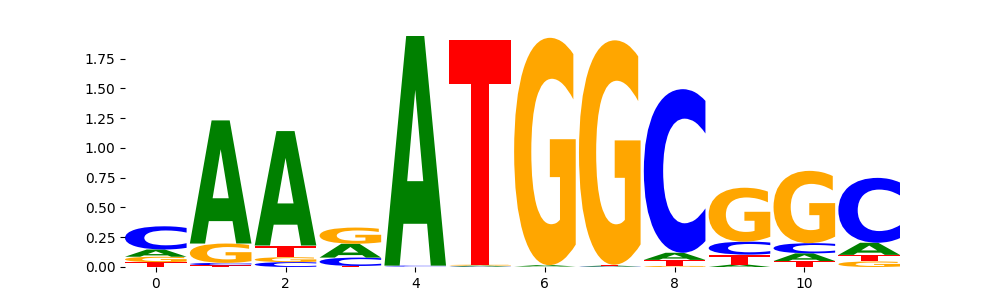 |
TYY1_HUMAN.H11MO.0.A | 7.161930e-06 |  |
YY1_MA0095.2 | 2.100540e-05 |  |
| pos__28 | 106 | 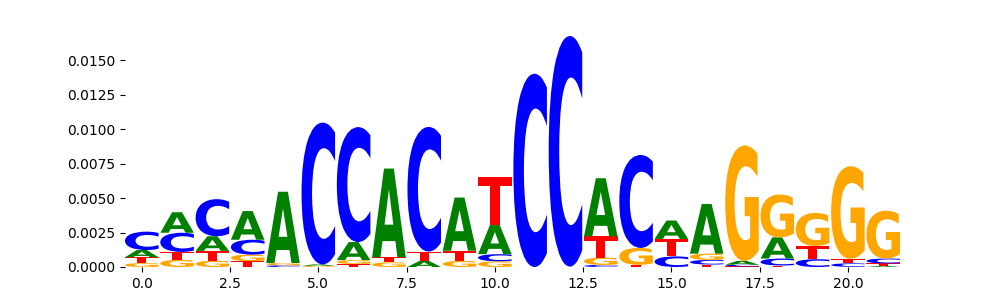 |
 |
SPI1_MOUSE.H11MO.0.A | 2.140450e-01 |  |
KLF4_MA0039.3 | 2.140450e-01 | 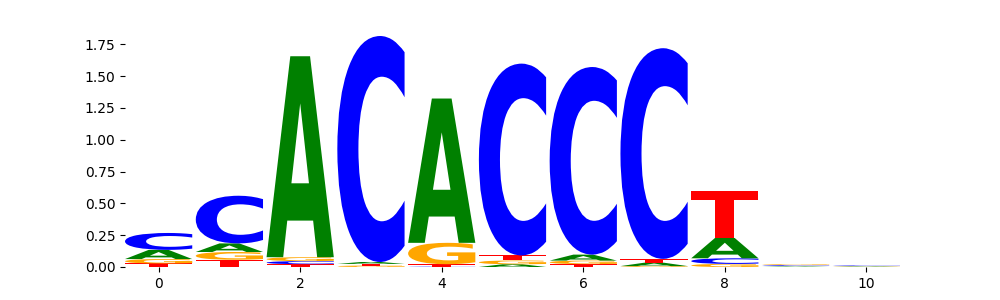 |
ELF5_HUMAN.H11MO.0.A | 3.850990e-01 | 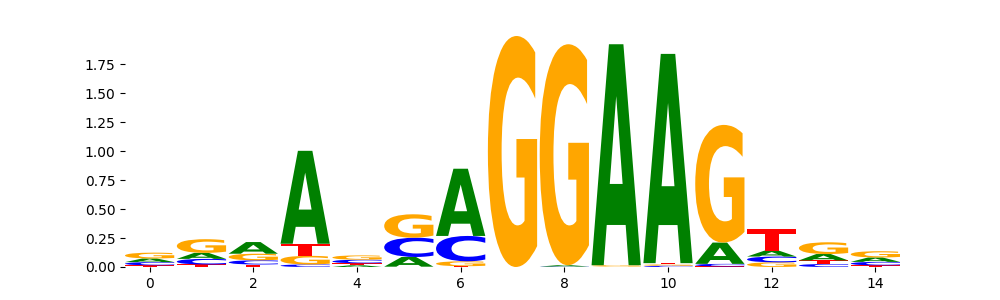 |
| pos__29 | 99 | 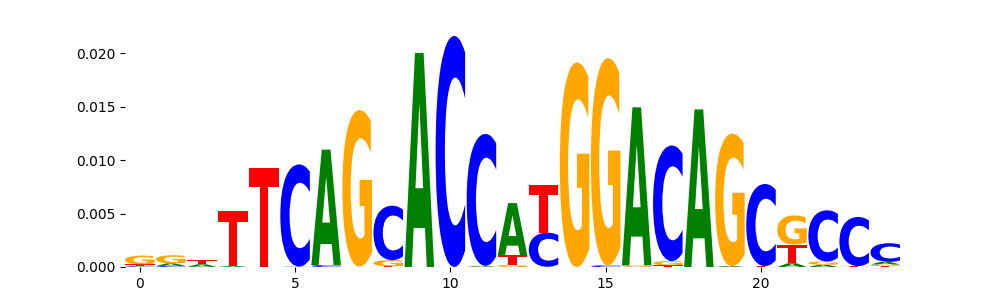 |
 |
REST_HUMAN.H11MO.0.A | 1.552590e-13 | 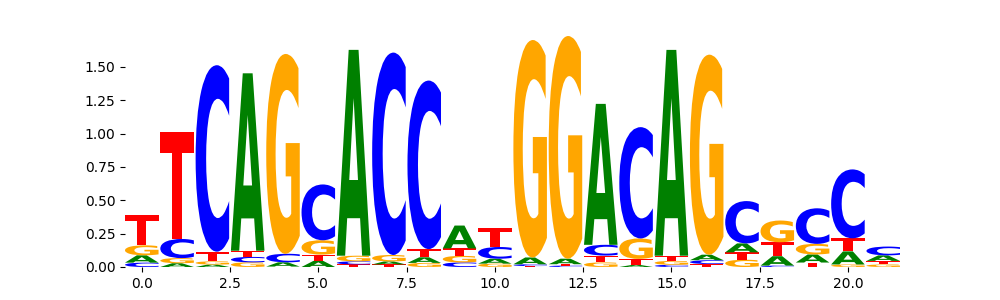 |
REST_MOUSE.H11MO.0.A | 5.767260e-12 | 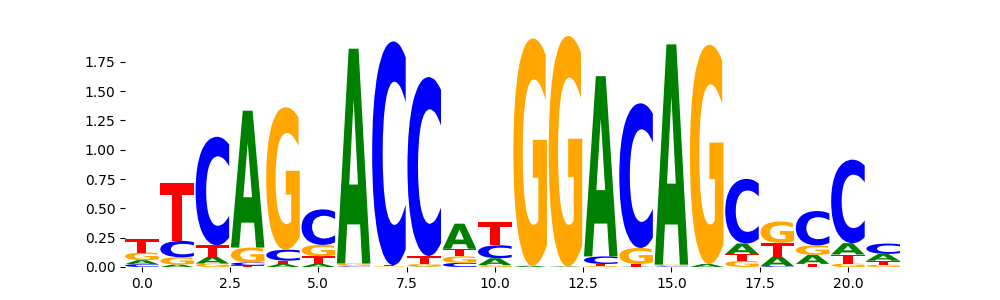 |
REST_MA0138.2 | 2.756050e-11 | 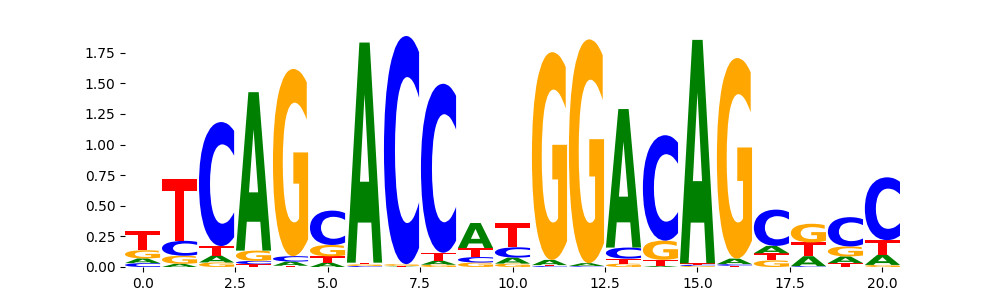 |
| pos__30 | 80 |  |
 |
RUNX3_RUNX_1 | 4.430910e-02 |  |
RUNX2_RUNX_1 | 4.430910e-02 |  |
RUNX1_MA0002.2 | 4.121630e-01 |  |
| pos__31 | 79 | 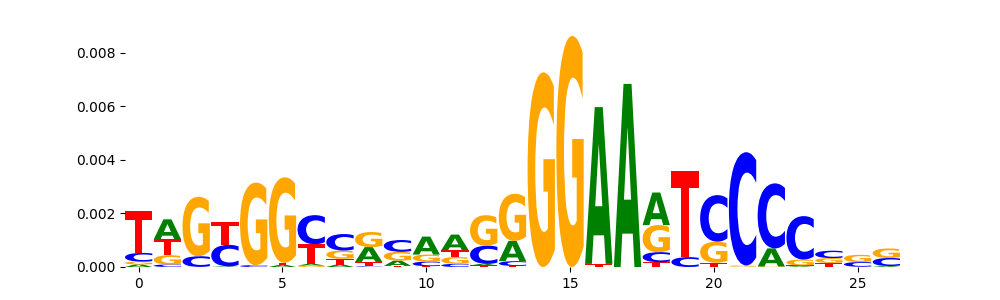 |
 |
NFKB2_HUMAN.H11MO.0.B | 1.300200e-02 | 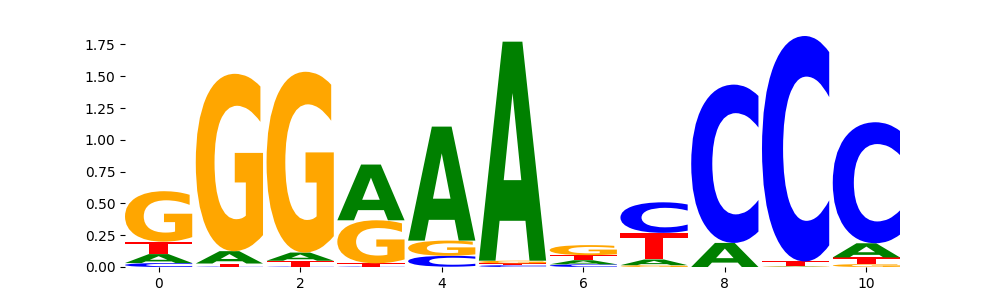 |
NFKB2_MOUSE.H11MO.0.C | 1.300200e-02 | 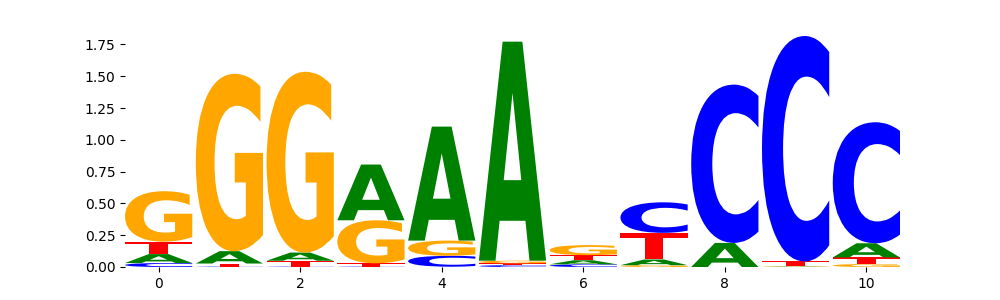 |
NFKB1_HUMAN.H11MO.1.B | 4.000100e-02 | 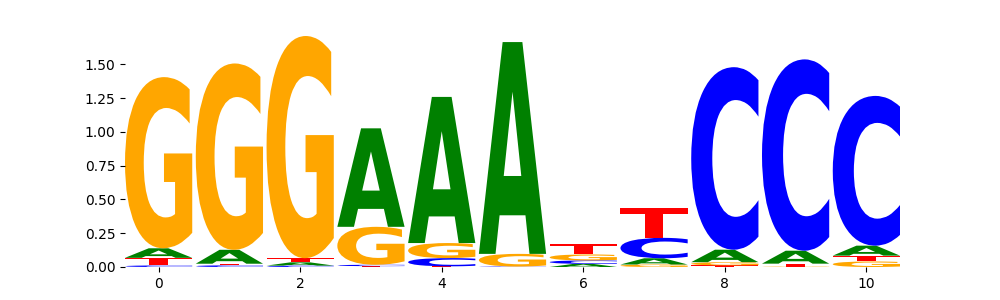 |
| pos__32 | 39 | 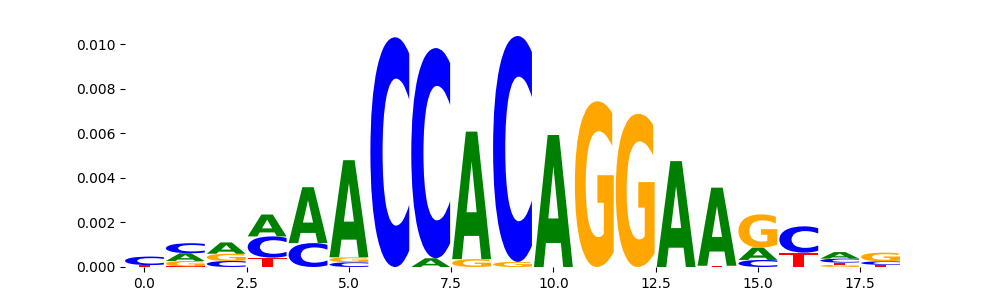 |
 |
RUNX2_HUMAN.H11MO.0.A | 4.872740e-01 |  |
RUNX2_MOUSE.H11MO.0.A | 4.872740e-01 |  |
RUNX1_MA0002.2 | 4.872740e-01 | 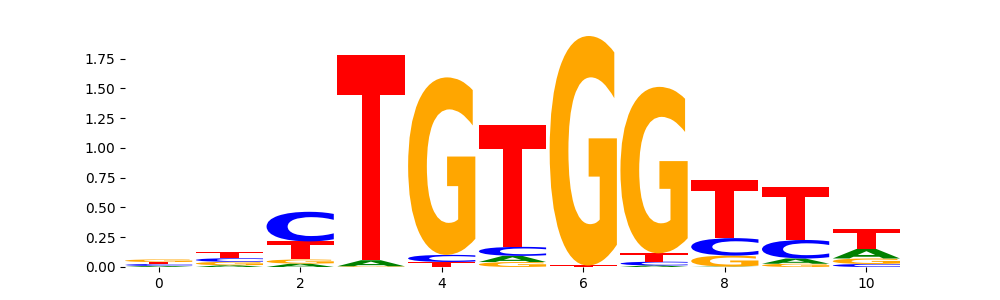 |
| pos__33 | 39 | 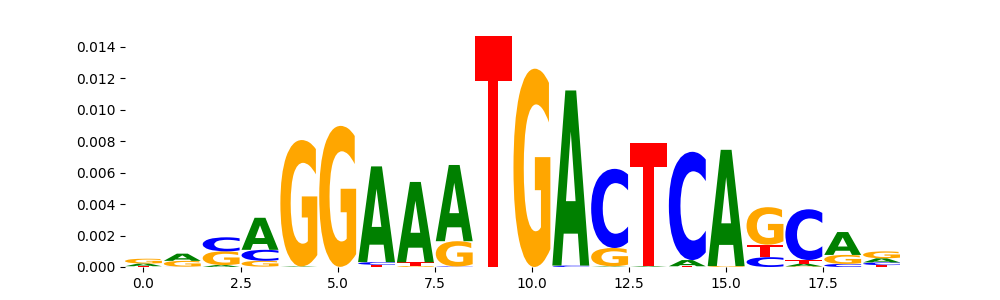 |
 |
JUN_MA0489.1 | 3.083330e-03 |  |
BATF+JUN_MA0462.1 | 3.083330e-03 |  |
FOSB_HUMAN.H11MO.0.A | 4.653980e-02 |  |
| pos__34 | 28 |  |
 |
TCF7_HUMAN.H11MO.0.A | 4.490890e-01 |  |
SOX2_HUMAN.H11MO.0.A | 4.490890e-01 | 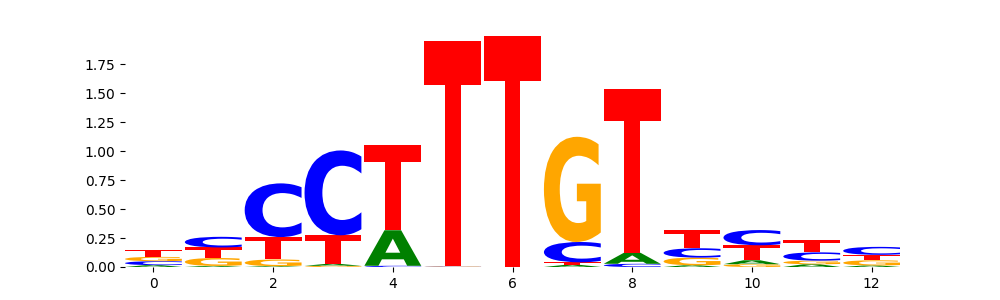 |
RUNX3_HUMAN.H11MO.0.A | 4.490890e-01 | 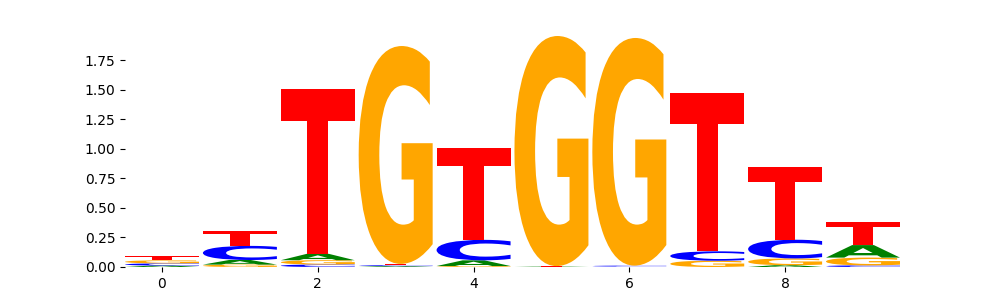 |
| pos__35 | 28 |  |
 |
HXA9_HUMAN.H11MO.0.B | 2.072160e-04 |  |
HXA9_MOUSE.H11MO.0.B | 2.072160e-04 |  |
MEIS1_HUMAN.H11MO.0.A | 2.072160e-04 | 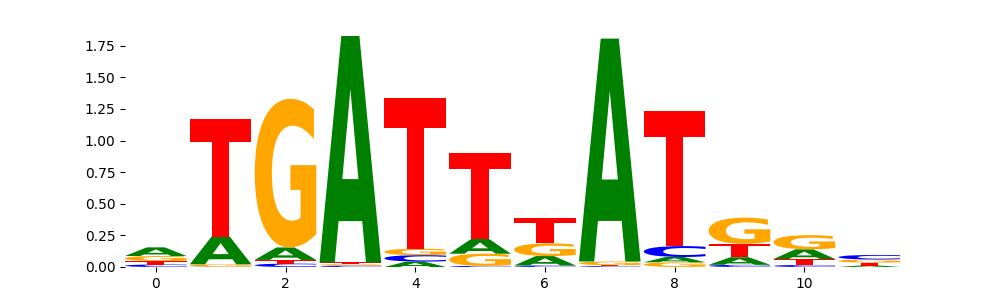 |
| pos__36 | 25 | 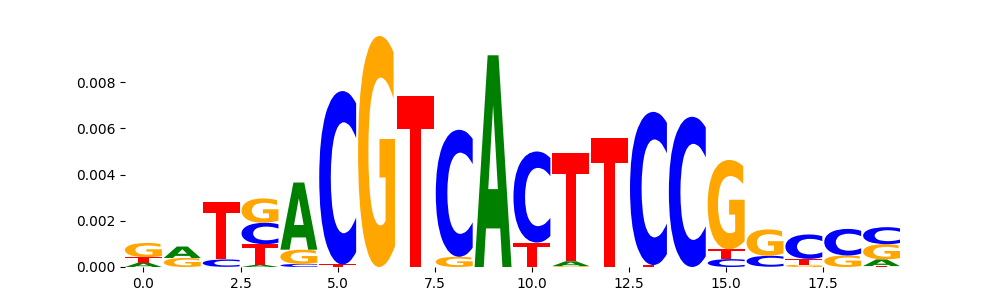 |
 |
Gabpa_MA0062.2 | 2.040150e-01 |  |
GMEB2_SAND_3 | 2.258190e-01 |  |
ELK1_HUMAN.H11MO.0.B | 2.258190e-01 | 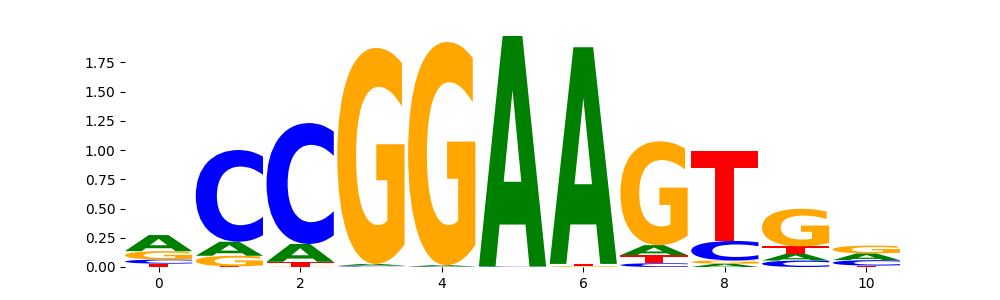 |
| pos__37 | 22 | 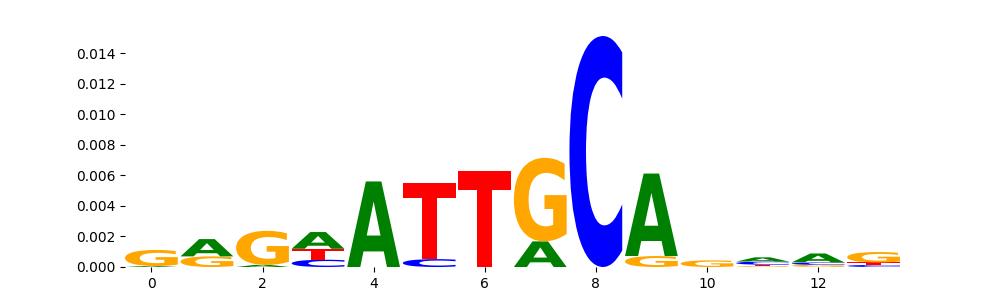 |
 |
Lhx8.mouse_homeodomain_3 | 1.990650e-01 | 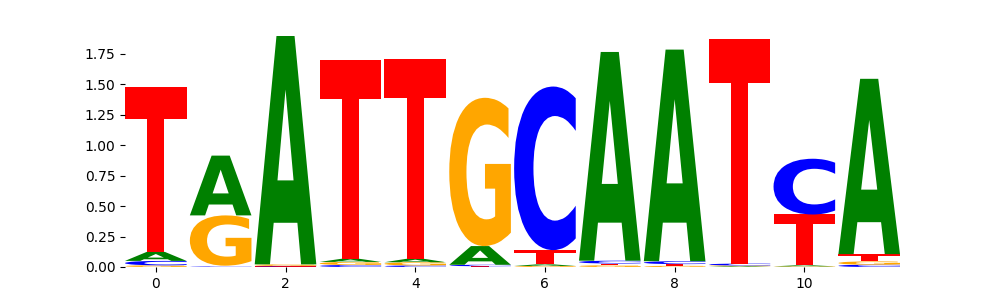 |
LHX6_homeodomain_3 | 1.990650e-01 | 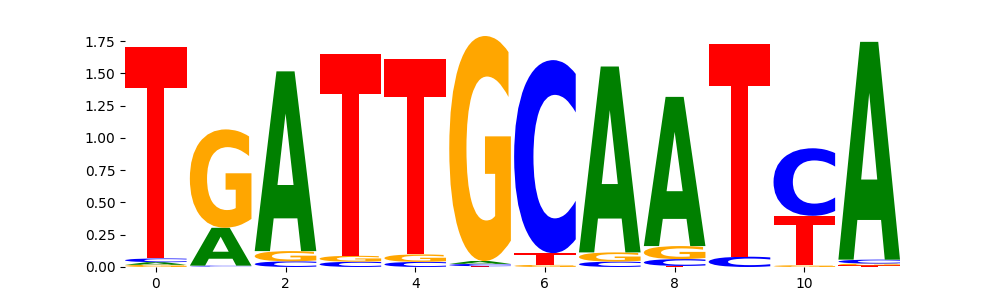 |
Ddit3+Cebpa_MA0019.1 | 1.990650e-01 | 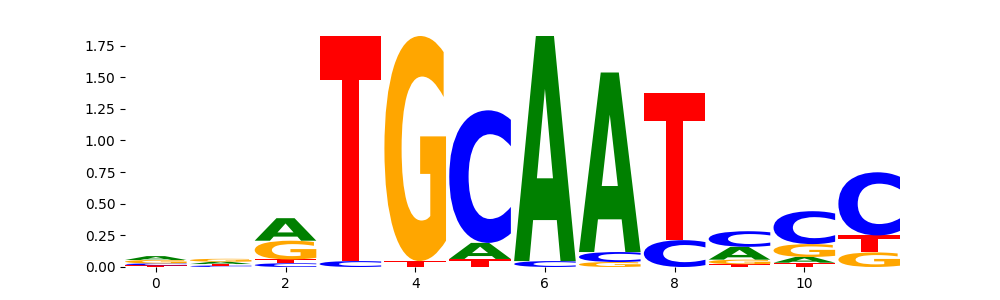 |
| pos__38 | 20 |  |
 |
ERG_HUMAN.H11MO.0.A | 1.355350e-01 |  |
ETS1_HUMAN.H11MO.0.A | 1.355350e-01 |  |
CTCF_MA0139.1 | 1.860400e-01 |  |
| neg__0 | 907 | 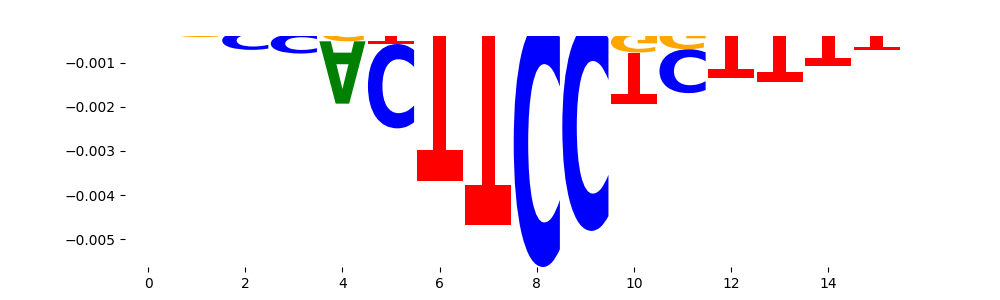 |
 |
ELF5_HUMAN.H11MO.0.A | 1.273570e-03 |  |
ELF3_HUMAN.H11MO.0.A | 2.960260e-03 |  |
ELF3_MOUSE.H11MO.0.B | 2.960260e-03 |  |
| neg__1 | 200 | 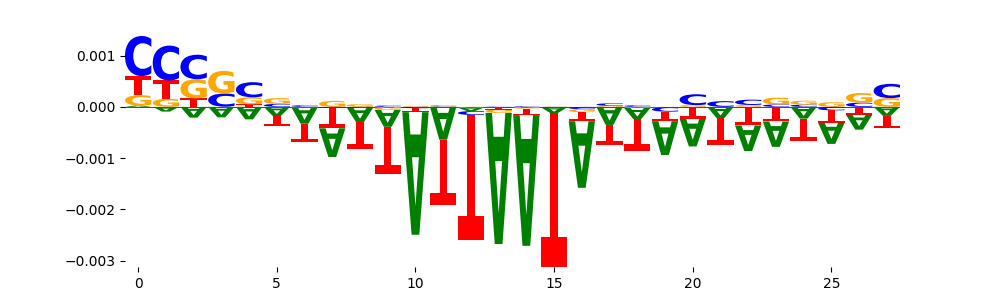 |
 |
DNASE_2 | 9.561450e-02 | 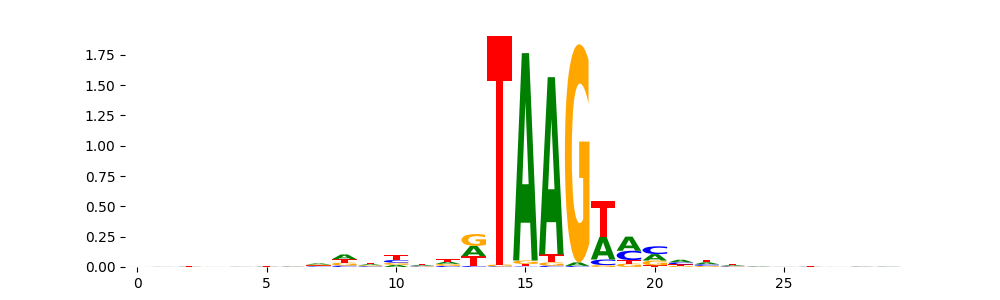 |
Arid3b_MA0601.1 | 9.561450e-02 |  |
FOXD2_forkhead_1 | 1.876450e-01 | 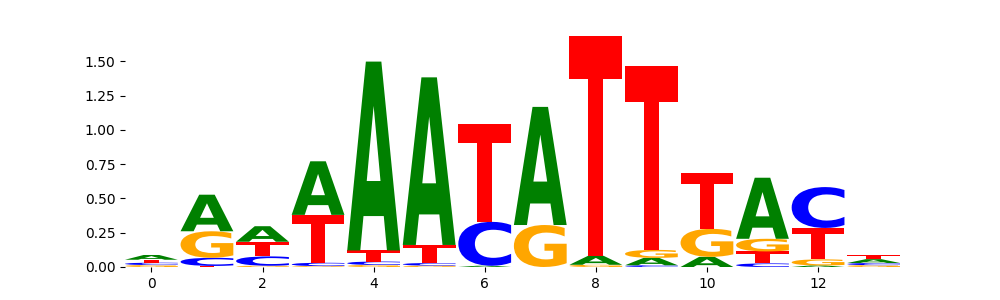 |
| neg__2 | 183 |  |
 |
RUNX3_HUMAN.H11MO.0.A | 1.403990e-02 | 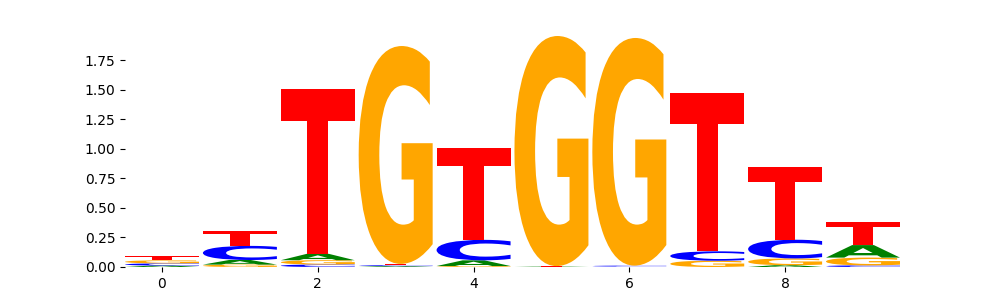 |
RUNX3_MOUSE.H11MO.0.A | 1.403990e-02 |  |
RUNX1_HUMAN.H11MO.0.A | 7.544920e-02 |  |
| neg__3 | 163 | 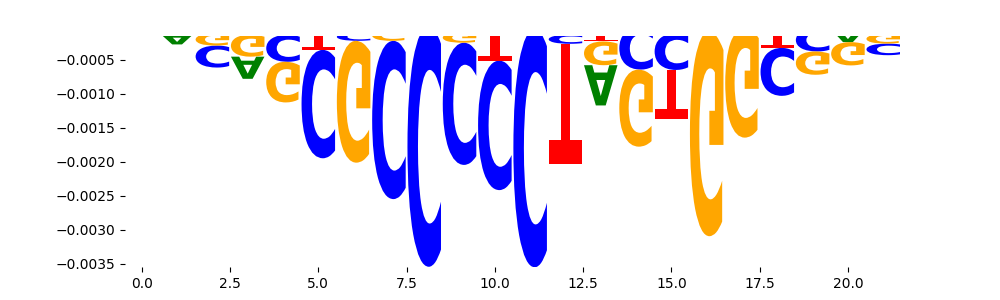 |
 |
CTCF_MA0139.1 | 9.969200e-09 |  |
CTCF_MOUSE.H11MO.0.A | 9.969200e-09 |  |
CTCFL_HUMAN.H11MO.0.A | 2.067740e-08 | 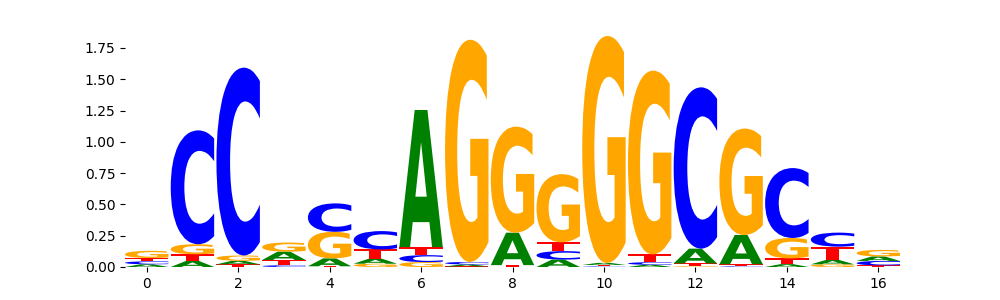 |
| neg__4 | 147 |  |
 |
SP3_HUMAN.H11MO.0.B | 6.191330e-06 |  |
SP3_MOUSE.H11MO.0.B | 6.191330e-06 |  |
SP1_MA0079.3 | 1.024550e-05 | 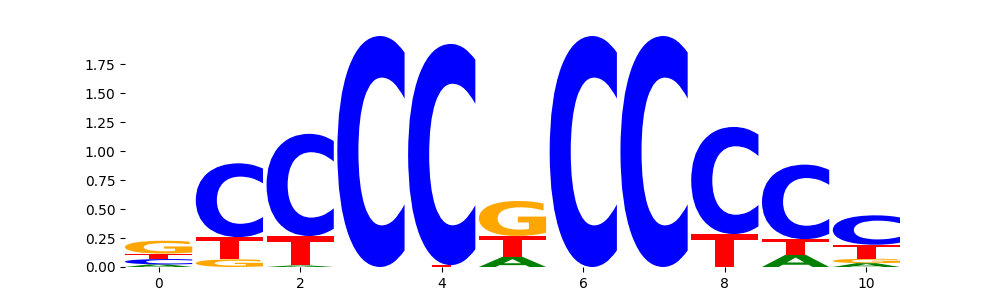 |
| neg__5 | 136 | 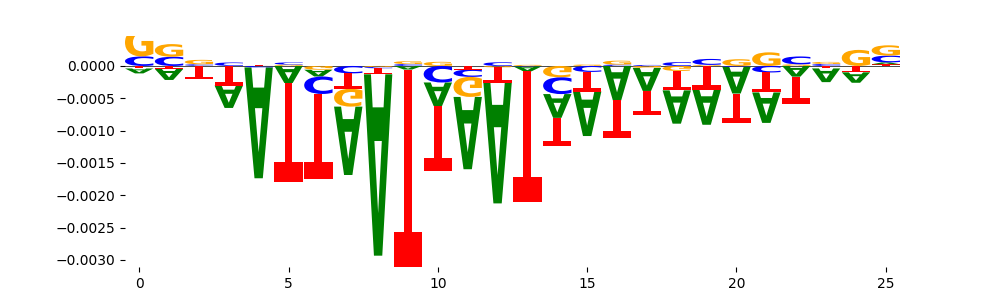 |
 |
LHX3_HUMAN.H11MO.0.C | 1.462540e-03 | 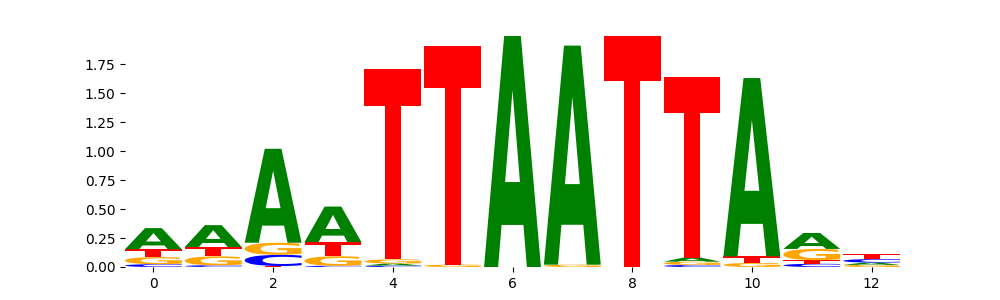 |
Lhx3_MA0135.1 | 2.234320e-03 | 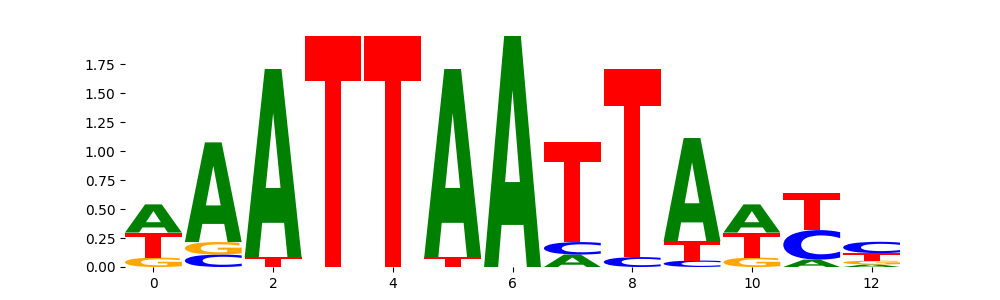 |
Hoxd8_MA0910.1 | 8.838770e-03 |  |
| neg__6 | 112 |  |
 |
CEBPB_MOUSE.H11MO.0.A | 5.979410e-07 |  |
CEBPA_MA0102.3 | 7.023930e-07 |  |
CEBPB_HUMAN.H11MO.0.A | 7.023930e-07 |  |
| neg__7 | 95 | 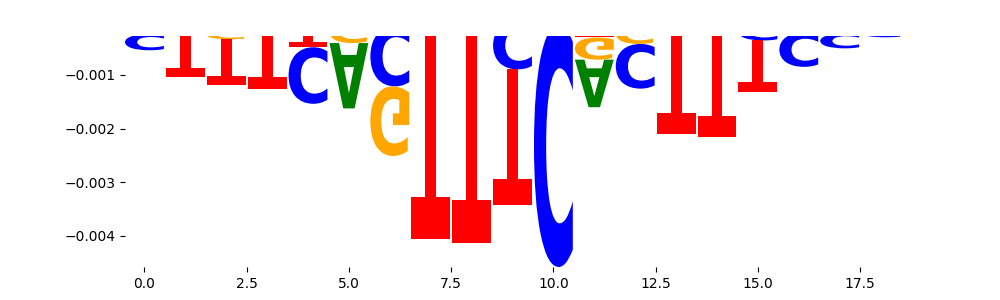 |
 |
IRF8_HUMAN.H11MO.0.B | 4.118100e-09 | 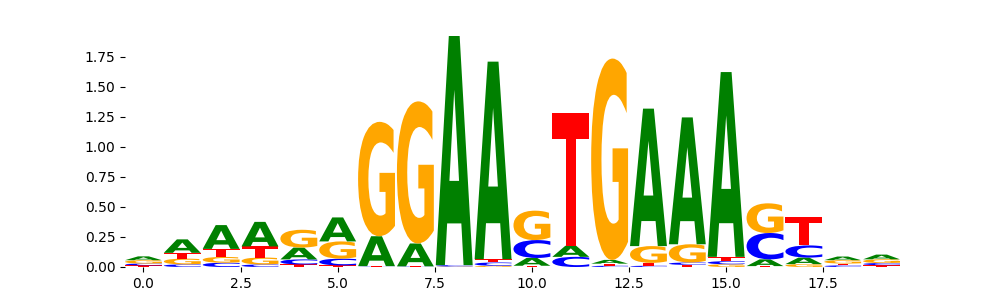 |
IRF8_MOUSE.H11MO.0.A | 4.118100e-09 | 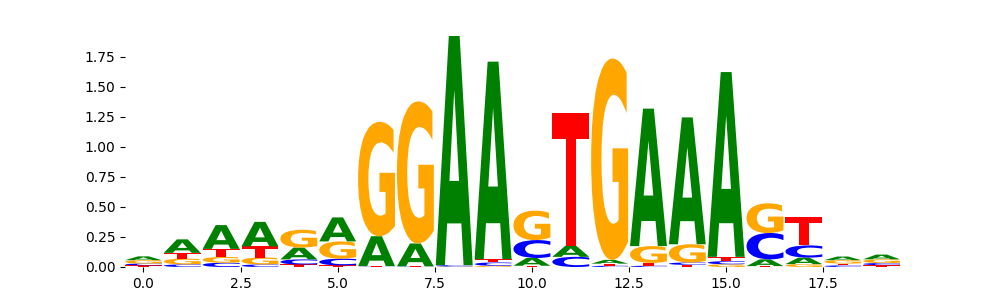 |
IRF1_MOUSE.H11MO.0.A | 1.030720e-07 |  |
| neg__8 | 88 | 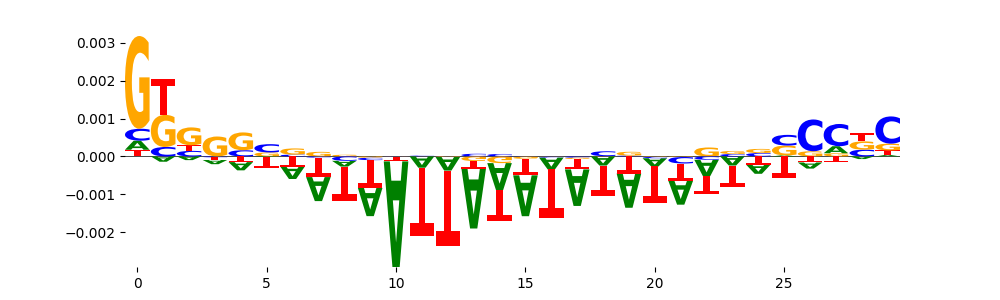 |
 |
DNASE_2 | 5.592540e-03 |  |
PIT1_HUMAN.H11MO.0.C | 2.661980e-01 |  |
PIT1_MOUSE.H11MO.0.C | 2.661980e-01 |  |
| neg__9 | 88 |  |
 |
FOXD2_forkhead_1 | 2.590780e-01 |  |
POU4F1_MA0790.1 | 2.590780e-01 |  |
POU4F1_POU_1 | 2.590780e-01 | 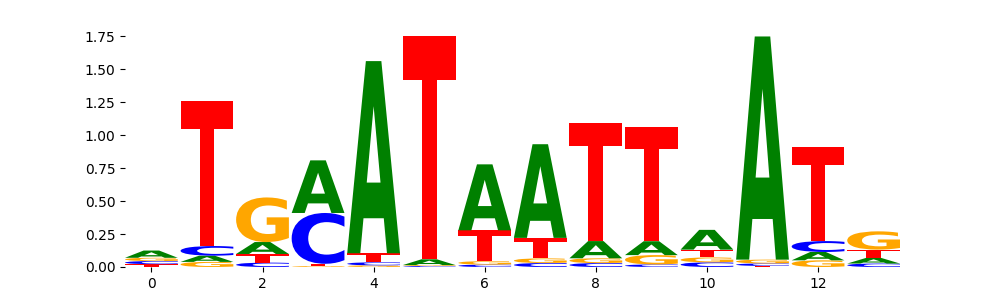 |
| neg__10 | 68 | 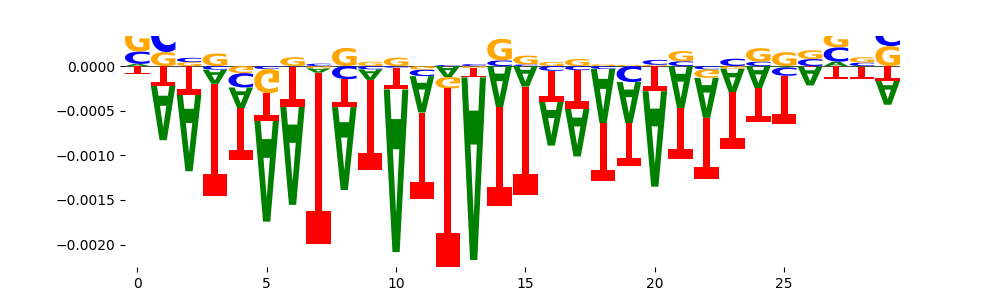 |
 |
POU3F3_POU_2 | 5.196520e-01 | 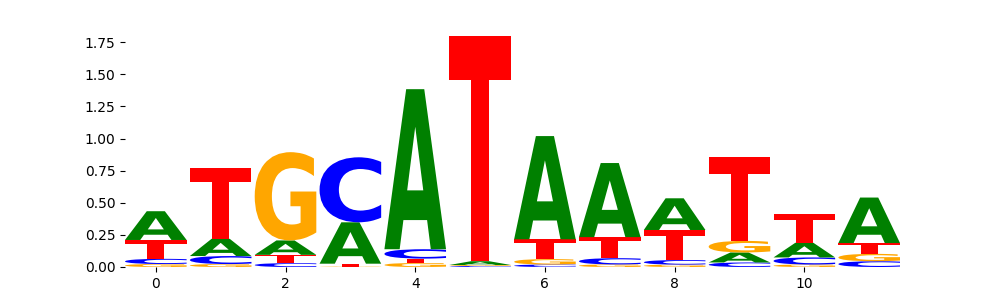 |
POU3F1_POU_2 | 5.196520e-01 | 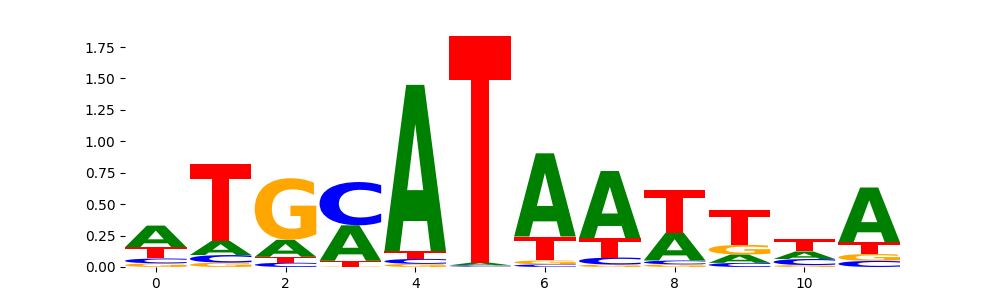 |
POU3F2_POU_1 | 5.196520e-01 | 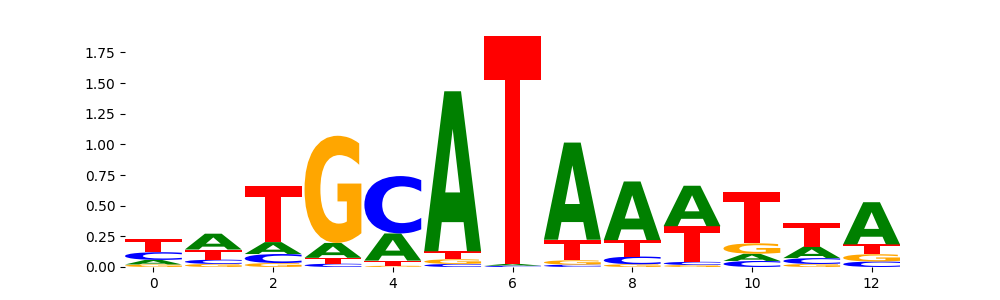 |
| neg__11 | 53 | 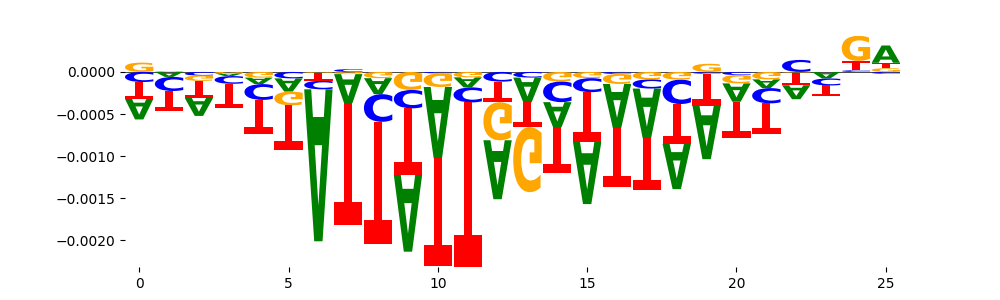 |
 |
MEF2D_MA0773.1 | 8.022430e-02 | 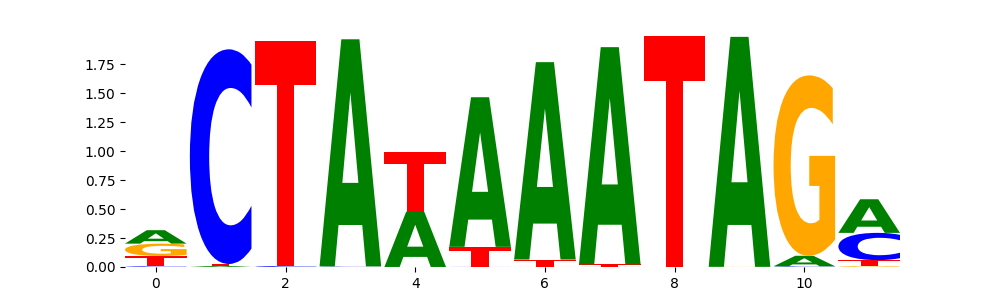 |
MEF2D_MADS_1 | 8.022430e-02 |  |
MEF2C_MA0497.1 | 8.022430e-02 | 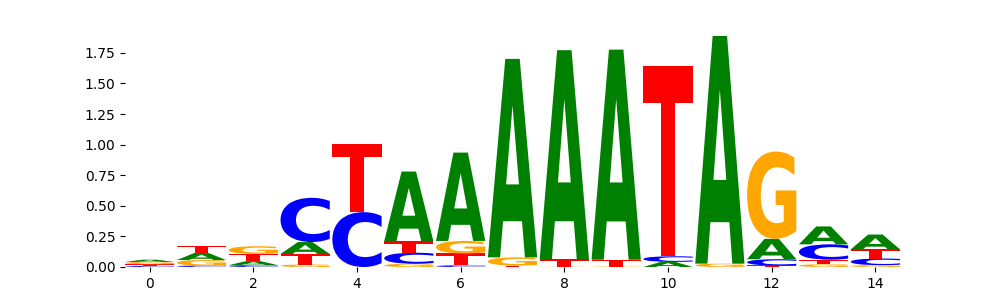 |
| neg__12 | 43 | 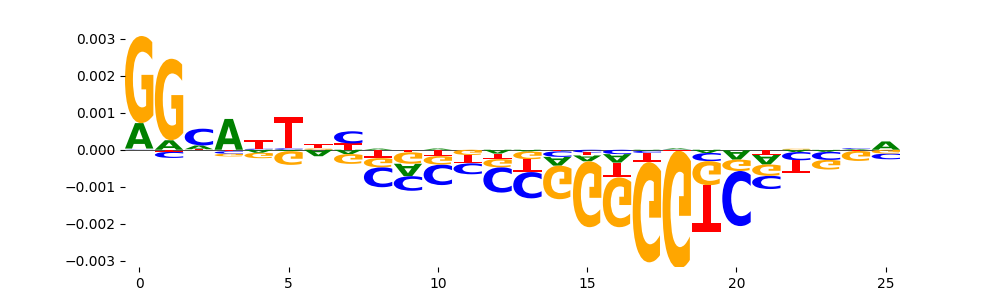 |
 |
ZIC1_HUMAN.H11MO.0.B | 1.511210e-01 | 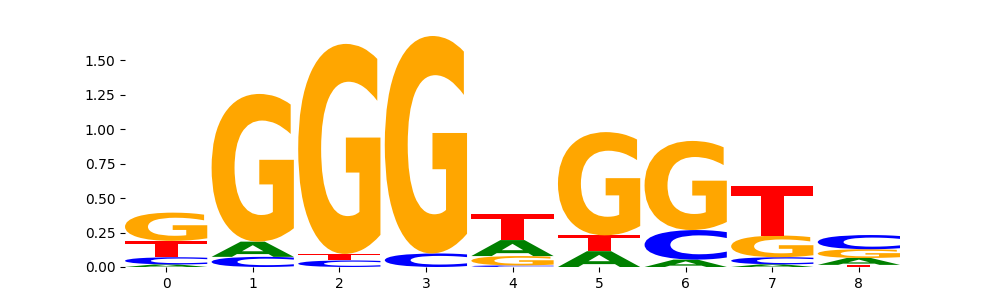 |
ZIC1_MOUSE.H11MO.0.B | 1.511210e-01 | 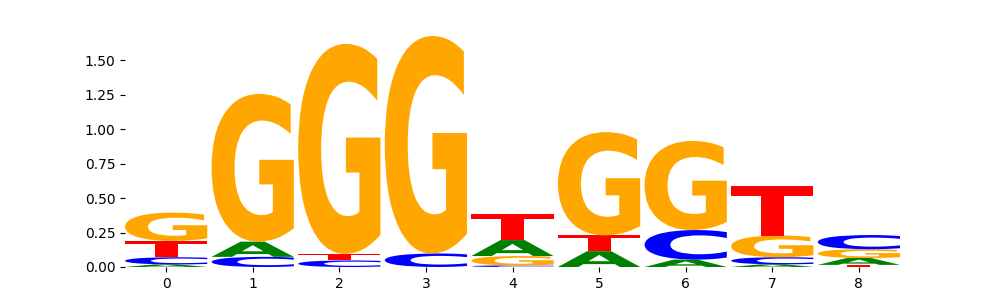 |
ZIC3_C2H2_1 | 1.511210e-01 | 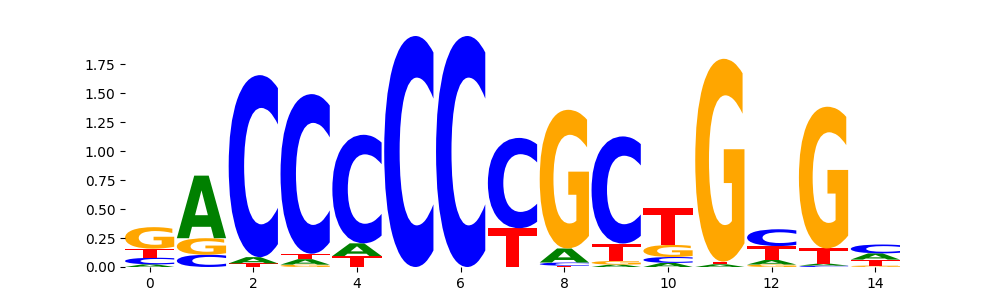 |
| neg__13 | 41 | 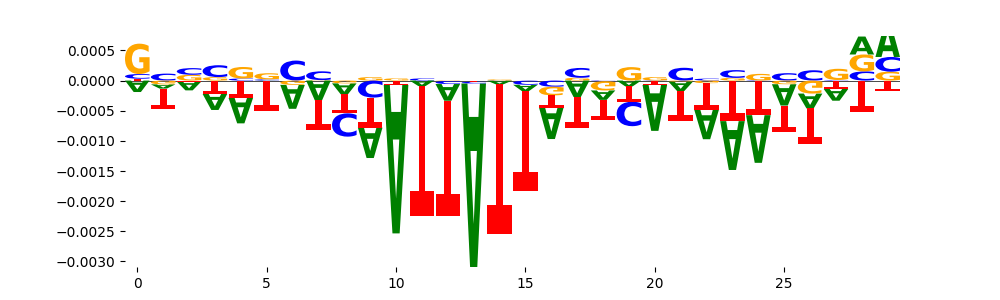 |
 |
CUX2_CUT_1 | 6.561920e-01 | 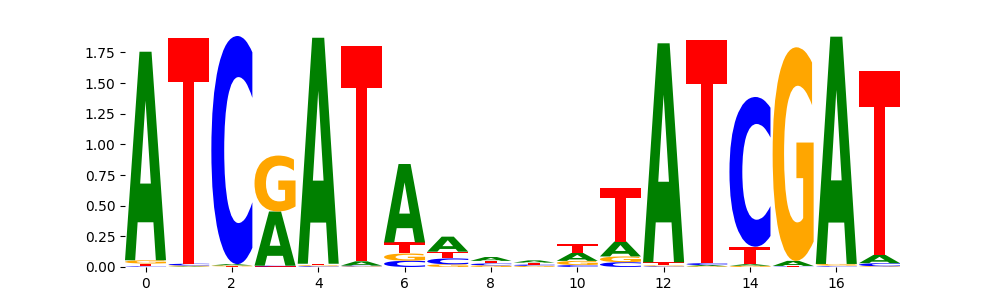 |
FOXG1_forkhead_1 | 6.561920e-01 |  |
ZNF384_MA1125.1 | 6.988890e-01 |  |
| neg__14 | 36 |  |
 |
NFIC_HUMAN.H11MO.0.A | 5.420420e-02 |  |
MXI1_HUMAN.H11MO.0.A | 1.624110e-01 | 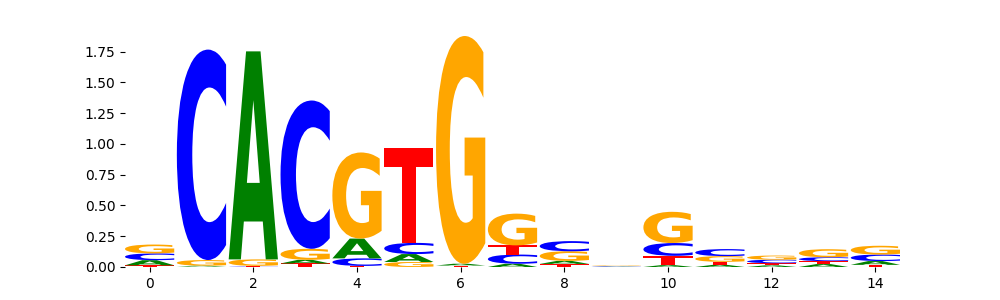 |
MXI1_MOUSE.H11MO.0.A | 1.624110e-01 | 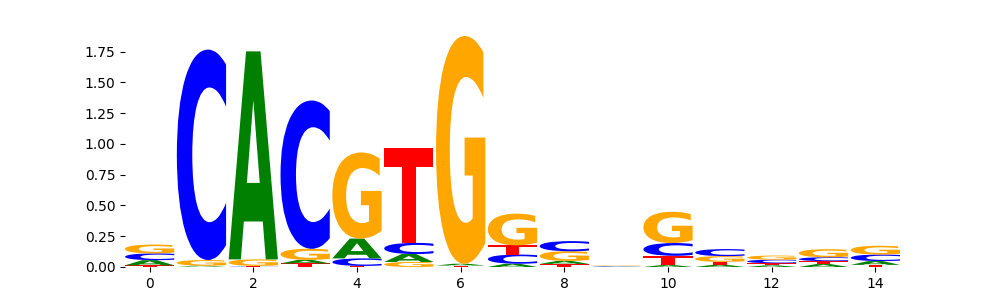 |
| neg__15 | 31 |  |
 |
EWSR1-FLI1_MA0149.1 | 8.923250e-03 |  |
FLI1_HUMAN.H11MO.0.A | 3.422790e-01 |  |
ETS2_HUMAN.H11MO.0.B | 4.636750e-01 |  |