Dup QC (all)
Flagstat QC (all, filtered)
PBC QC (all)
| Total Read Pairs | Distinct Read Pairs | One Read Pair | Two Read Pairs | NRF = Distinct/Total | PBC1 = OnePair/Distinct | PBC2 = OnePair/TwoPair | |
|---|---|---|---|---|---|---|---|
| rep1 | 103059191 | 81619699 | 65132263 | 13095793 | 0.791969 | 0.797997 | 4.973526 |
| rep2 | 44295404 | 38354958 | 33398886 | 4310013 | 0.865890 | 0.870784 | 7.749138 |
NRF (non redundant fraction)
PBC1 (PCR Bottleneck coefficient 1)
PBC2 (PCR Bottleneck coefficient 2)
PBC1 is the primary measure. Provisionally
- 0-0.5 is severe bottlenecking
- 0.5-0.8 is moderate bottlenecking
- 0.8-0.9 is mild bottlenecking
- 0.9-1.0 is no bottlenecking
Cross-correlation QC (all)
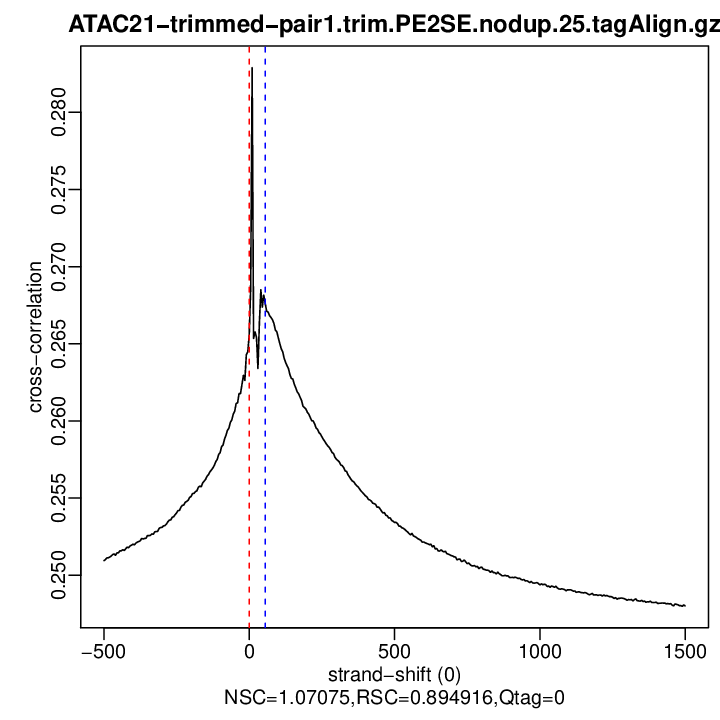
rep1 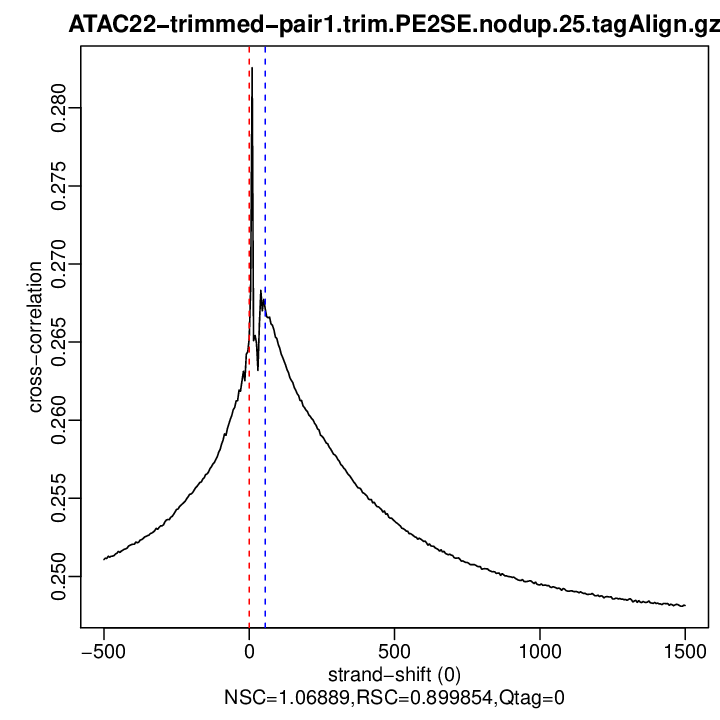
rep2
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | phantomPeakCoef | relPhantomPeakCoef | QualityTag | |
|---|---|---|---|---|---|---|---|---|---|---|
| rep1 | 25000000 | 0 | 0.265560858672526 | 55 | 0.2676213 | 1500 | 0.2480135 | 1.070752 | 0.8949163 | 0 |
| rep2 | 25000000 | 0 | 0.265230395730548 | 55 | 0.2671327 | 1500 | 0.2481371 | 1.068886 | 0.8998537 | 0 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display


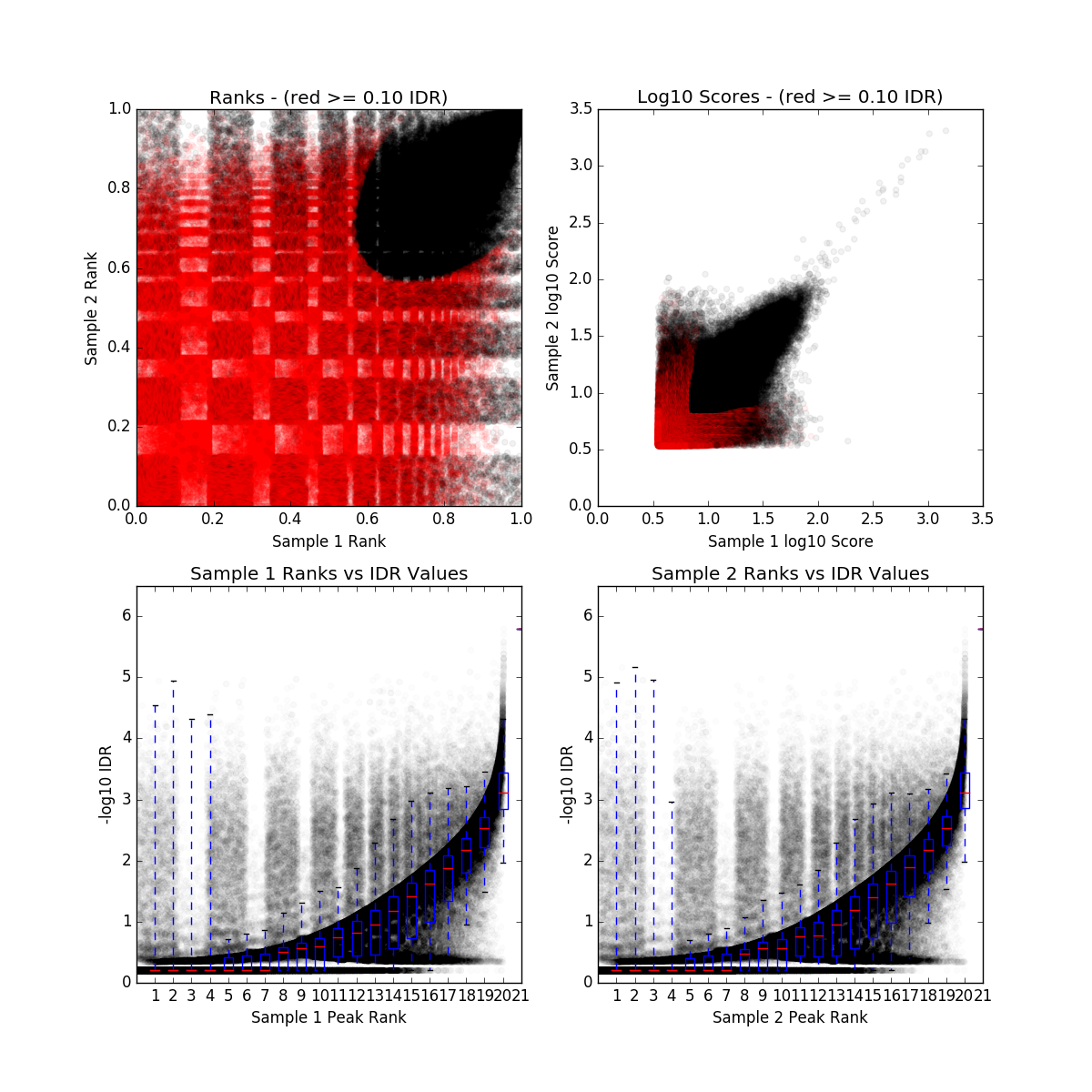
IDR QC (idr)
IDR_final.qc
| Nt | Np | conservative_set | optimal_set | rescue_ratio | self_consistency_ratio | reproducibility |
|---|---|---|---|---|---|---|
| 83545 | 0 | rep1-rep2 | rep1-rep2 | Infinity | NaN | 0 |