Bowtie2 QC (all)
| Total Reads | % Aligned | |
|---|---|---|
| rep1 | 62689806 | 98.45 |
Dup QC (all)
| Unpaired Reads | Paired Reads | Unmapped Reads | Unpaired Dupes | Paired Dupes | Paired Opt. Dupes | % Dupes | |
|---|---|---|---|---|---|---|---|
| rep1 | 0 | 43000932 | 0 | 0 | 30479056 | 25938130 | 0.7088 |
Flagstat QC (all, filtered)
| Reads (QC-passed) | Reads (QC-failed) | Dupes (QC-passed) | Dupes (QC-failed) | Mapped Reads | % Mapped | |
|---|---|---|---|---|---|---|
| rep1 | 25043752 | 0 | 0 | 0 | 25043752 | 100.00 |
PBC QC (all)
| Total Read Pairs | Distinct Read Pairs | One Read Pair | Two Read Pairs | NRF = Distinct/Total | PBC1 = OnePair/Distinct | PBC2 = OnePair/TwoPair | |
|---|---|---|---|---|---|---|---|
| rep1 | 11164796 | 10127760 | 9319956 | 752490 | 0.907116 | 0.920239 | 12.385488 |
NRF (non redundant fraction)
PBC1 (PCR Bottleneck coefficient 1)
PBC2 (PCR Bottleneck coefficient 2)
PBC1 is the primary measure. Provisionally
- 0-0.5 is severe bottlenecking
- 0.5-0.8 is moderate bottlenecking
- 0.8-0.9 is mild bottlenecking
- 0.9-1.0 is no bottlenecking
Cross-correlation QC (all)
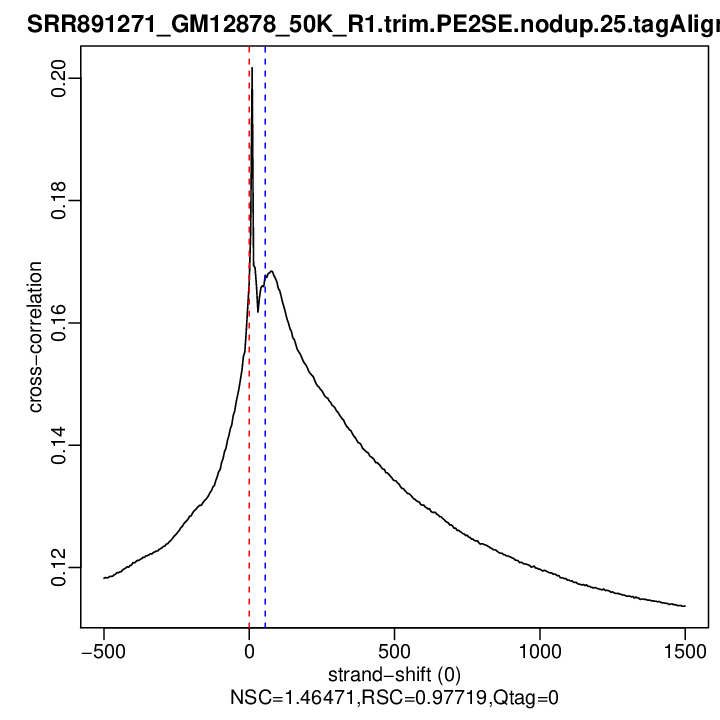
rep1
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | phantomPeakCoef | relPhantomPeakCoef | QualityTag | |
|---|---|---|---|---|---|---|---|---|---|---|
| rep1 | 10127760 | 0 | 0.166541540326861 | 55 | 0.1677749 | 1500 | 0.113703 | 1.464706 | 0.9771897 | 0 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display
