![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | GD01-trimmed-pair1.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 37450600 |
| Filtered Sequences | 0 |
| Sequence length | 21-150 |
| %GC | 43 |
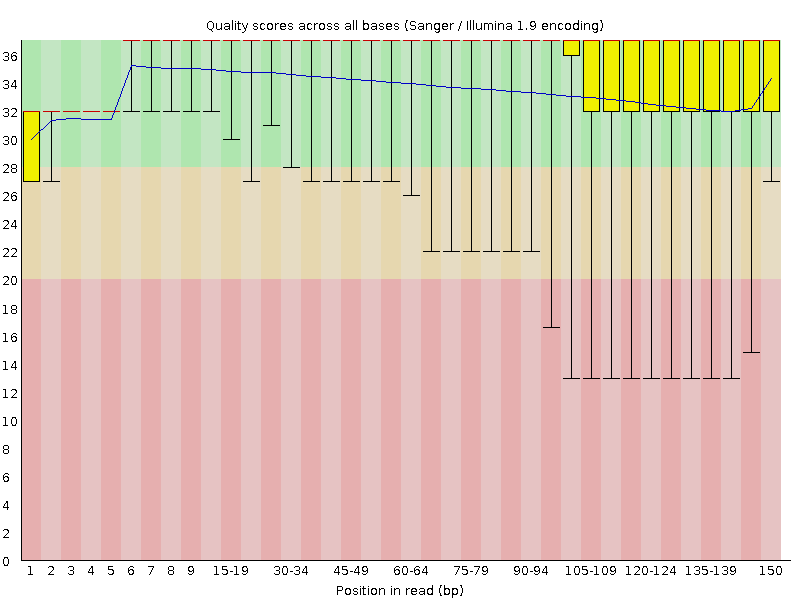
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
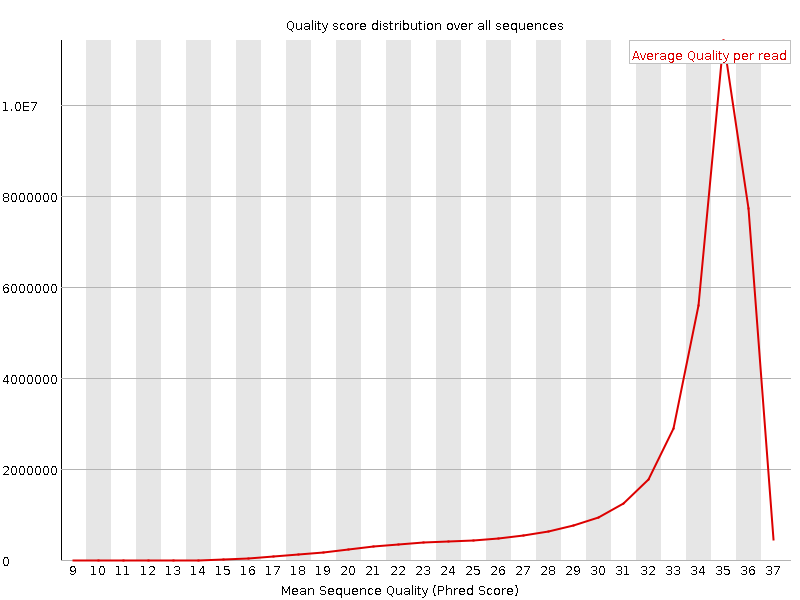
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
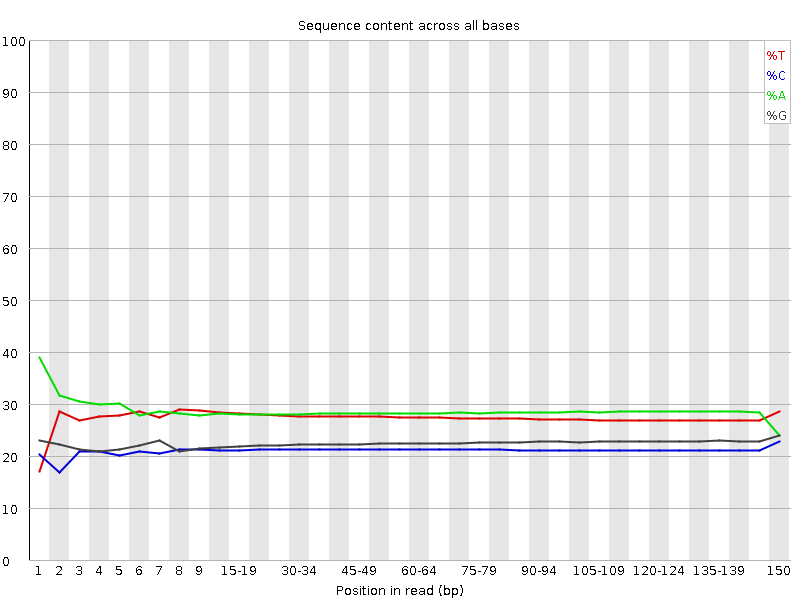
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
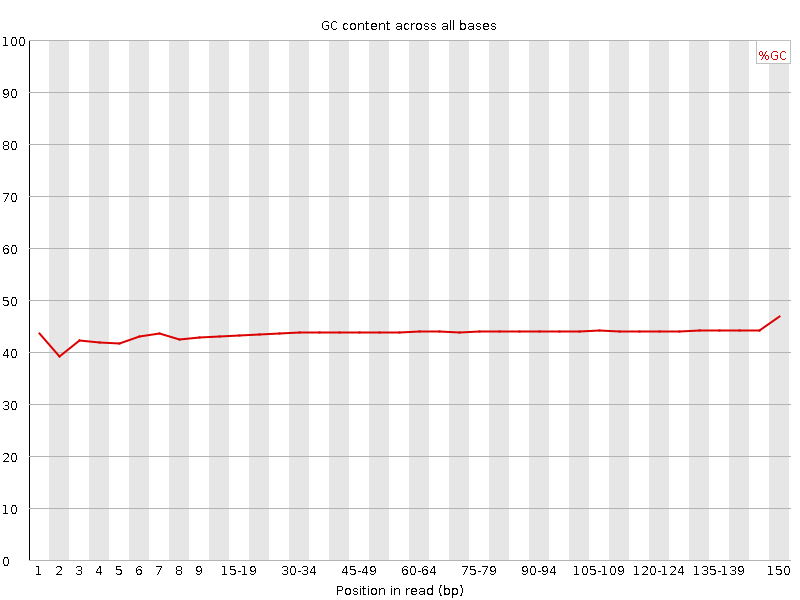
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
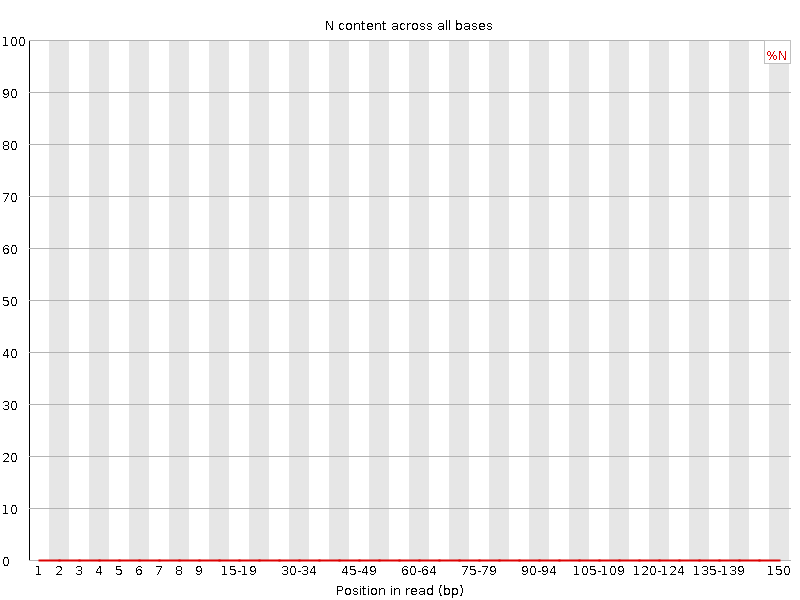
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
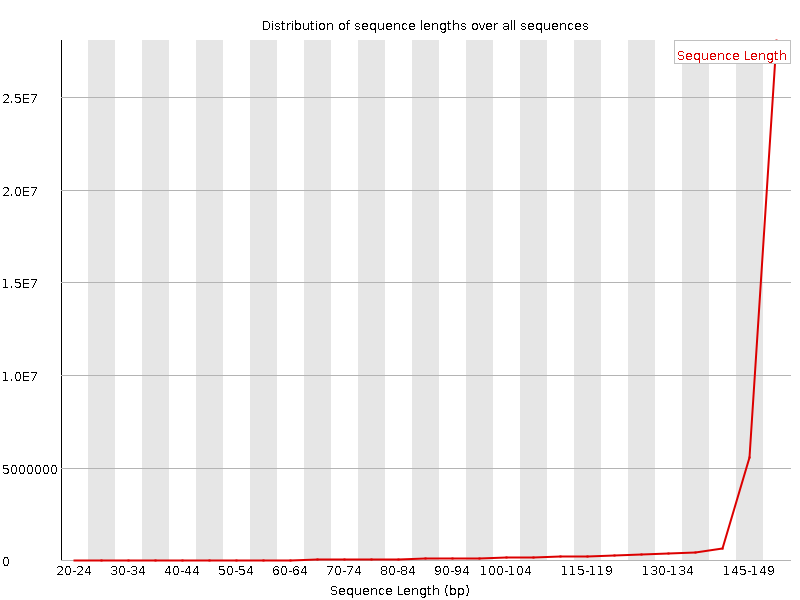
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 51612 | 0.13781354637842919 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
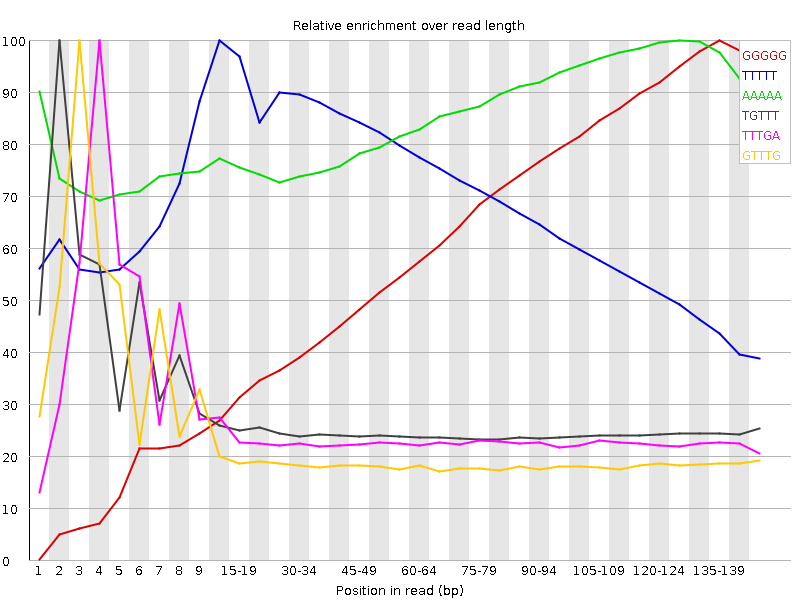
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 35963280 | 11.334372 | 18.07204 | 135-139 |
| TTTTT | 50527525 | 6.069385 | 8.671839 | 10-14 |
| AAAAA | 40972685 | 4.017237 | 4.6722207 | 125-129 |
| TGTTT | 10070110 | 1.467017 | 5.6868305 | 2 |
| TTTGA | 9409805 | 1.3162696 | 5.4398274 | 4 |
| GTTTG | 7278160 | 1.2858988 | 6.404611 | 3 |
| GTGTT | 5733425 | 1.0129765 | 6.181457 | 1 |