![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | D2_25somitomere-p9c10r2-trimmed-pair1.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1830151 |
| Filtered Sequences | 0 |
| Sequence length | 88-151 |
| %GC | 47 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

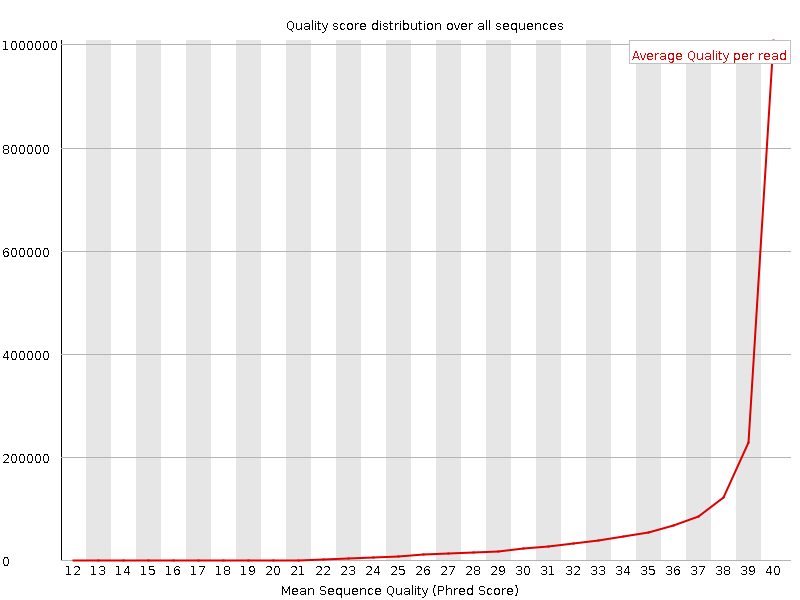
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
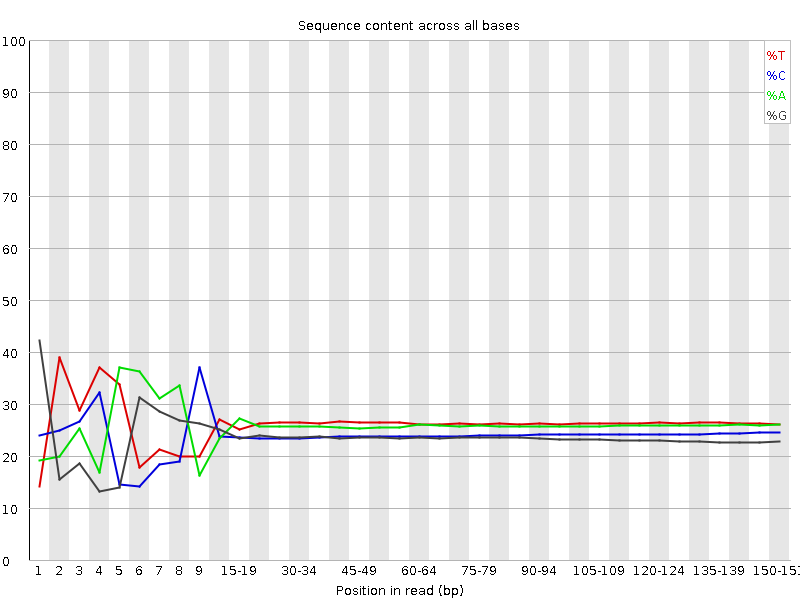
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
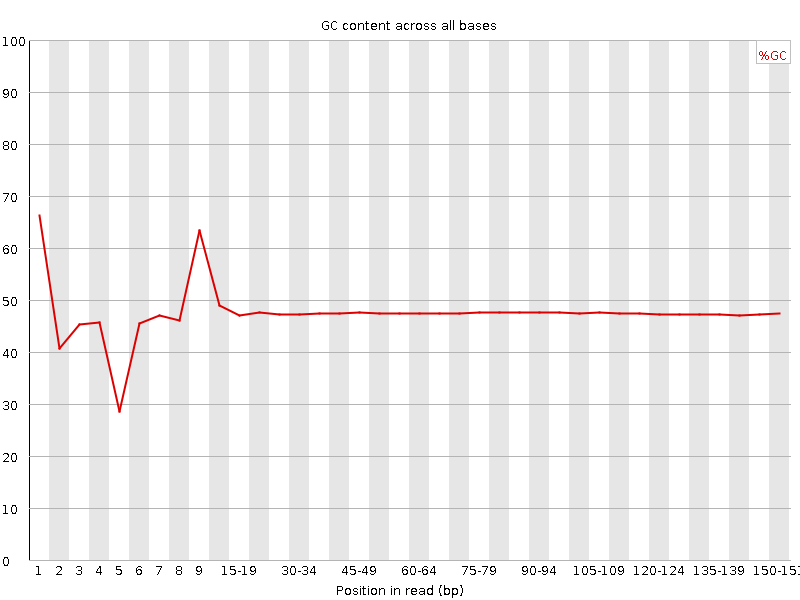
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content
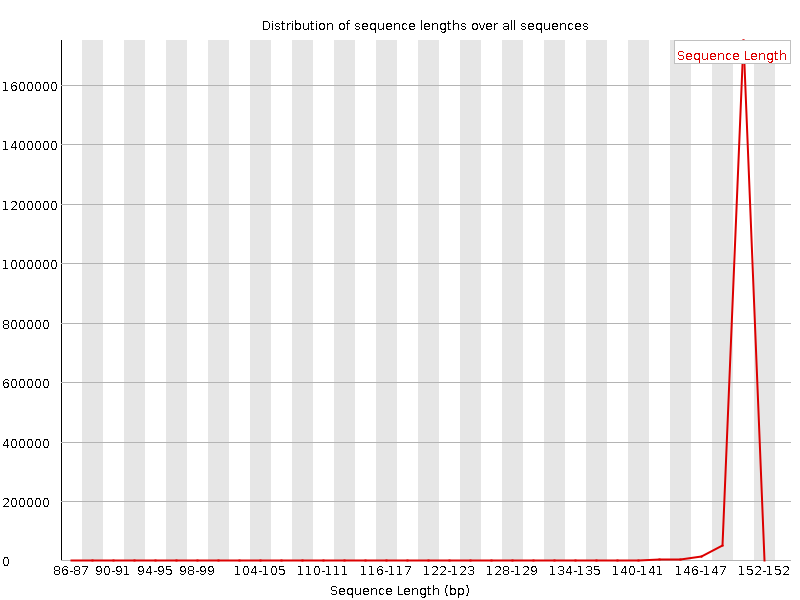
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
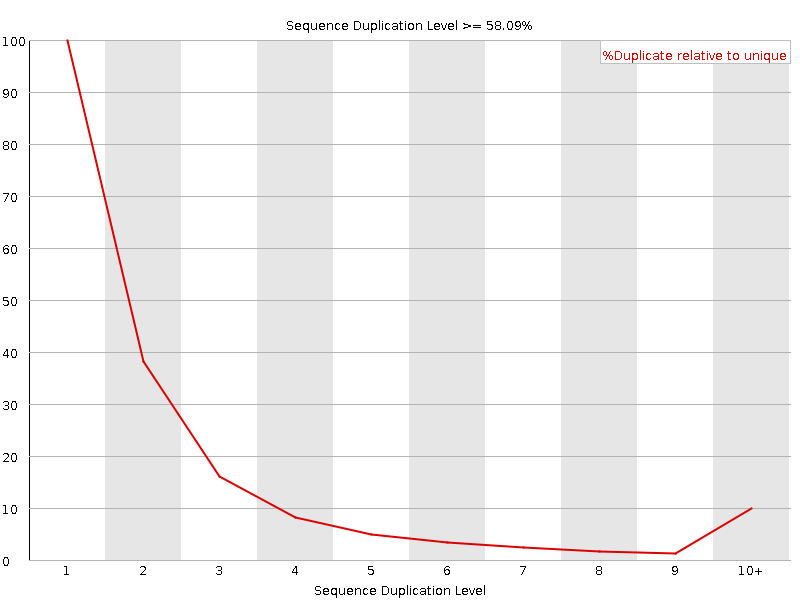
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTATACACATCTCCGAGCCCACGAGACGTAGAGGAATCTCGTATGCCGT | 4988 | 0.2725458172577017 | No Hit |
| GTATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 3011 | 0.16452194381775057 | No Hit |
| ATACACATCTCCGAGCCCACGAGACGTAGAGGAATCTCGTATGCCGTCTT | 2059 | 0.11250437805405127 | TruSeq Adapter, Index 3 (95% over 21bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content

| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 1850220 | 5.3500357 | 9.080539 | 25-29 |
| AAAAA | 1110795 | 3.5290625 | 7.187326 | 145-147 |
| CAGCA | 562095 | 2.2777817 | 5.101497 | 4 |
| CCAGG | 509795 | 2.2744796 | 7.649327 | 3 |
| CTGGG | 506585 | 2.2688107 | 5.71378 | 4 |
| CCCAG | 509540 | 2.2224243 | 6.341223 | 2 |
| AGGAG | 522385 | 2.2149746 | 5.70336 | 5 |
| GGCTG | 487815 | 2.1847467 | 5.9190626 | 7 |
| CCTGG | 498725 | 2.1835806 | 5.768892 | 3 |
| AGAAG | 563910 | 2.1717193 | 5.0530963 | 5 |
| CTCCT | 550600 | 2.1006021 | 5.067379 | 1 |
| CAGAG | 498560 | 2.0666058 | 11.002382 | 4 |
| TCCTG | 528130 | 2.0610394 | 6.7167964 | 2 |
| ACACA | 559020 | 2.057527 | 8.657062 | 6 |
| CAGGA | 486665 | 2.0172994 | 7.125092 | 4 |
| CAGGC | 445980 | 1.9897658 | 5.0392165 | 9 |
| TGGAG | 471650 | 1.9625443 | 5.566444 | 5 |
| CTGGA | 462965 | 1.8832585 | 5.887987 | 4 |
| TCCAG | 464355 | 1.8466052 | 6.707243 | 2 |
| CACAG | 450495 | 1.8255441 | 6.2056174 | 7 |
| GCAGA | 440210 | 1.8247365 | 10.88376 | 9 |
| CATGG | 441255 | 1.7949461 | 15.703288 | 4 |
| TGGGG | 390495 | 1.7889546 | 10.590669 | 6 |
| CTGTG | 437430 | 1.7461916 | 6.8539433 | 9 |
| ACATC | 482430 | 1.742505 | 5.072951 | 8 |
| GGGAG | 372460 | 1.7387692 | 5.1357718 | 7 |
| CACTG | 435770 | 1.732931 | 5.999436 | 7 |
| CAGGG | 376490 | 1.7182165 | 5.9261913 | 4 |
| CAGTG | 419115 | 1.7048845 | 5.817711 | 9 |
| ATACA | 449215 | 1.5074664 | 5.261092 | 4 |
| ACAGA | 398530 | 1.5004346 | 5.7130594 | 8 |
| ACAGC | 357810 | 1.4499562 | 5.195125 | 8 |
| ATGGG | 347225 | 1.4448097 | 14.959818 | 5 |
| TCTTG | 399665 | 1.4220563 | 5.4290686 | 2 |
| TACAC | 393570 | 1.4215486 | 8.27039 | 5 |
| AACAG | 361300 | 1.3602662 | 5.138715 | 7 |
| GTGTG | 326570 | 1.3335137 | 5.4297266 | 1 |
| ACATG | 360330 | 1.3313065 | 13.815615 | 3 |
| GACAG | 307535 | 1.2747786 | 5.843133 | 7 |
| GTCTT | 351805 | 1.2517647 | 5.626725 | 1 |
| GGGGG | 233765 | 1.201506 | 5.379261 | 7 |
| ACTGT | 328170 | 1.189866 | 5.8449264 | 8 |
| AGAGT | 307320 | 1.1614654 | 7.101465 | 5 |
| ATCAA | 344550 | 1.1562338 | 9.094279 | 3 |
| GGACA | 276870 | 1.1476676 | 5.0071106 | 6 |
| ACAGT | 299580 | 1.1068542 | 5.893714 | 8 |
| GTCCT | 276705 | 1.0798477 | 5.126044 | 1 |
| TCAAC | 289695 | 1.0463591 | 9.563229 | 4 |
| GTCCA | 258315 | 1.0272439 | 5.4605217 | 1 |
| GAGTA | 262230 | 0.9910551 | 10.159297 | 1 |
| GTACA | 261090 | 0.96464574 | 17.161022 | 1 |
| AGTAC | 247150 | 0.9131418 | 7.645004 | 2 |
| TACAT | 270005 | 0.889174 | 12.632149 | 2 |
| TATCA | 244950 | 0.8066635 | 9.701413 | 2 |
| CGCAG | 178160 | 0.79487115 | 11.004381 | 8 |
| GTGTA | 184315 | 0.68359315 | 5.7531567 | 1 |
| GGTAT | 166055 | 0.6158698 | 6.642876 | 1 |
| GTATC | 160680 | 0.58258724 | 10.378805 | 1 |
| ACGCA | 126960 | 0.51448095 | 10.018744 | 7 |
| CAACG | 124780 | 0.5056469 | 10.003912 | 5 |
| GTATA | 138620 | 0.46695897 | 5.4708366 | 1 |
| AACGC | 110870 | 0.44927928 | 9.989051 | 6 |