Flagstat QC (all)
Dup QC (all)
Flagstat QC (all, filtered)
PBC QC (all)
| Total Read Pairs | Distinct Read Pairs | One Read Pair | Two Read Pairs | NRF = Distinct/Total | PBC1 = OnePair/Distinct | PBC2 = OnePair/TwoPair | |
|---|---|---|---|---|---|---|---|
| rep1 | 46253951 | 38462893 | 31911362 | 5512639 | 0.831559 | 0.829666 | 5.788763 |
| rep2 | 45295358 | 33979326 | 25220360 | 6740158 | 0.750172 | 0.742227 | 3.741805 |
| ctl1 | 49175935 | 44013622 | 39709076 | 3877910 | 0.895024 | 0.902200 | 10.239814 |
NRF (non redundant fraction)
PBC1 (PCR Bottleneck coefficient 1)
PBC2 (PCR Bottleneck coefficient 2)
PBC1 is the primary measure. Provisionally
- 0-0.5 is severe bottlenecking
- 0.5-0.8 is moderate bottlenecking
- 0.8-0.9 is mild bottlenecking
- 0.9-1.0 is no bottlenecking
Cross-correlation QC (all)

rep1 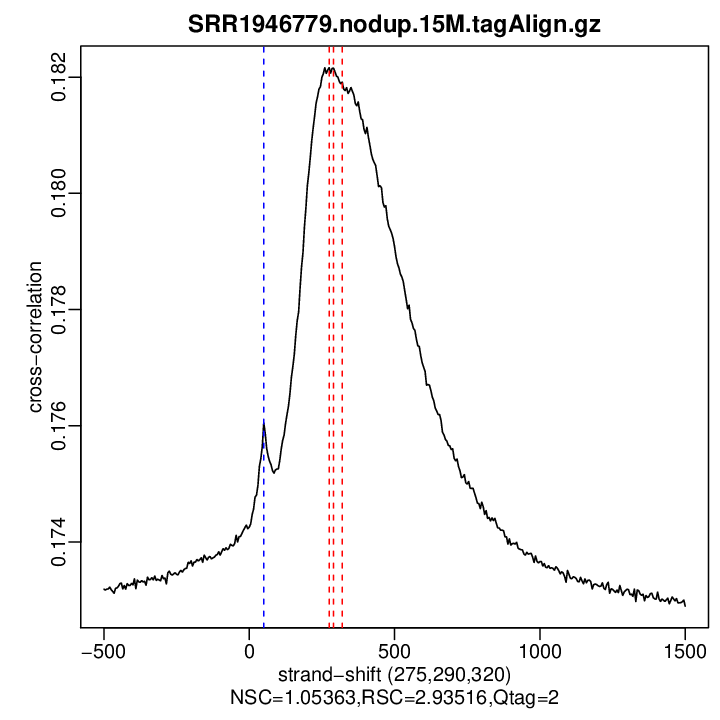
rep2
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | NSC | RSC | |
|---|---|---|---|---|---|---|---|---|---|
| rep1 | 15000000 | 255 | 0.178926314707944 | 50 | 0.175066 | 1500 | 0.1721764 | 1.039203 | 2.335939 |
| rep2 | 15000000 | 275 | 0.182164001894418 | 50 | 0.1760504 | 1500 | 0.1728912 | 1.053634 | 2.935156 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display

