Bowtie2 QC (all)
| Total Reads | % Aligned | |
|---|---|---|
| rep1 | 86508988 | 98.64 |
Dup QC (all)
| Unpaired Reads | Paired Reads | Unmapped Reads | Unpaired Dupes | Paired Dupes | Paired Opt. Dupes | % Dupes | |
|---|---|---|---|---|---|---|---|
| rep1 | 58417837 | 0 | 0 | 5141196 | 0 | 0 | 0.088007 |
Flagstat QC (all, filtered)
| Reads (QC-passed) | Reads (QC-failed) | Dupes (QC-passed) | Dupes (QC-failed) | Mapped Reads | % Mapped | |
|---|---|---|---|---|---|---|
| rep1 | 53276641 | 0 | 0 | 0 | 53276641 | 100.00 |
PBC QC (all)
| Total Read Pairs | Distinct Read Pairs | One Read Pair | Two Read Pairs | NRF = Distinct/Total | PBC1 = OnePair/Distinct | PBC2 = OnePair/TwoPair | |
|---|---|---|---|---|---|---|---|
| rep1 | 58201498 | 53262731 | 49894928 | 2603567 | 0.915144 | 0.936770 | 19.164065 |
NRF (non redundant fraction)
PBC1 (PCR Bottleneck coefficient 1)
PBC2 (PCR Bottleneck coefficient 2)
PBC1 is the primary measure. Provisionally
- 0-0.5 is severe bottlenecking
- 0.5-0.8 is moderate bottlenecking
- 0.8-0.9 is mild bottlenecking
- 0.9-1.0 is no bottlenecking
Cross-correlation QC (all)
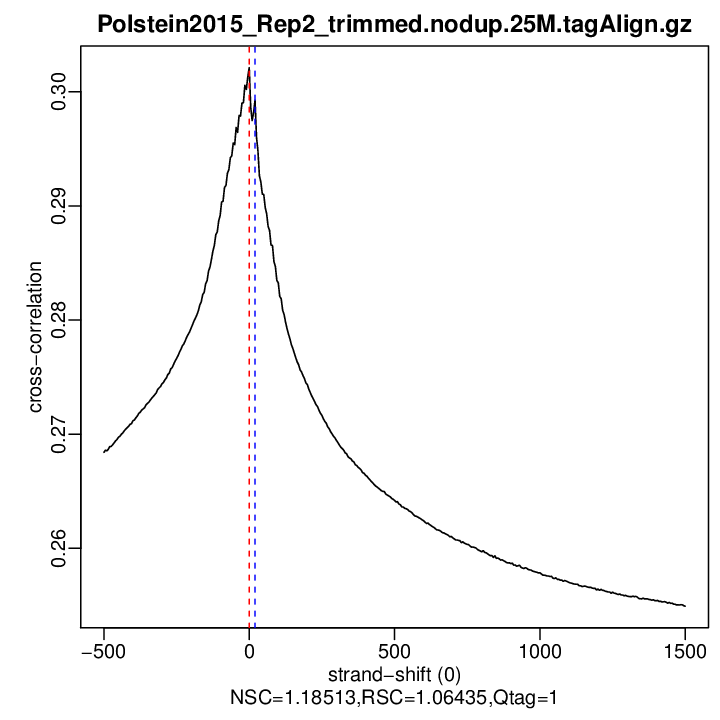
rep1
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | NSC | RSC | |
|---|---|---|---|---|---|---|---|---|---|
| rep1 | 25000000 | 0 | 0.302112422399815 | 20 | 0.2992592 | 1500 | 0.2549183 | 1.185134 | 1.064348 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display
