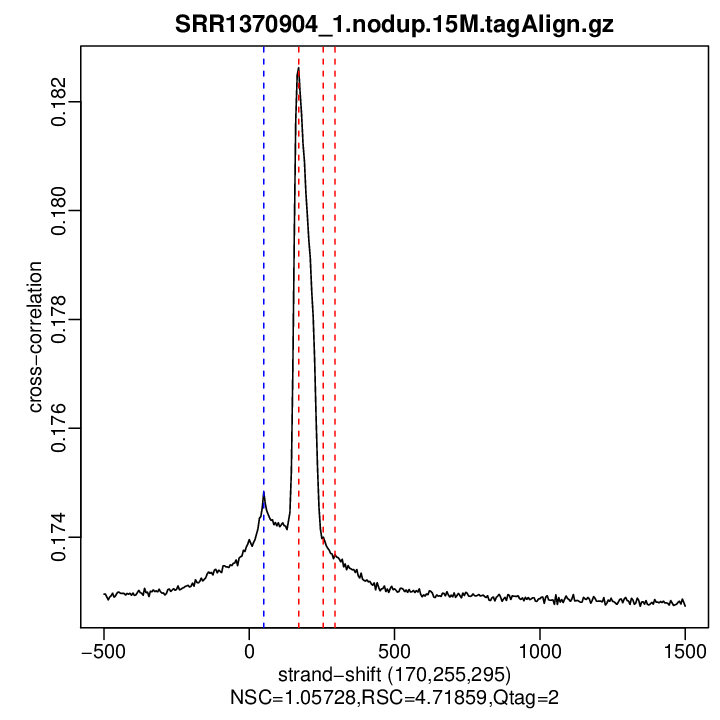
Enrichment QC (strand cross-correlation measures)
| Reads | 15000000 |
|---|---|
| Est. Fragment Len. | 170 |
| Corr. Est. Fragment Len. | 0.182626302070605 |
| Phantom Peak | 50 |
| Corr. Phantom Peak | 0.1748288 |
| Argmin. Corr. | 1500 |
| Min. Corr. | 0.1727319 |
| NSC | 1.057282 |
| RSC | 4.718593 |
NOTE: Reads from replicates are subsampled to a max of 15M
- Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
- Relative strand cross-correlation coefficient (RSC) = col10 in outFile
- Estimated fragment length = col3 in outFile, take the top value
Number of peaks
| rep1 | 300000 |
|---|---|
| rep1-pr1 | 284205 |
| rep1-pr2 | 284262 |
| overlap | 16340 |
- ppr1: Raw peaks called on the first pooled pseudoreplicates
- ppr2: Raw peaks called on the second pooled pseudoreplicates
- repi: Raw peaks called on true replicate i
- repi-pr1 : Raw peaks called on the first pseudoreplicate from replicate i
- repi-pr2 : Raw peaks called on the second pseudoreplicates from replicate i
- overlap : Overlapped peaks (filtered if blacklist exists)
Enrichment QC (Fraction of reads in peaks)
| Fraction of Reads in Peak | 0.000755717 |
|---|
- ppr: IDR peaks comparing pooled pseudo replicates
- rep1-pr: IDR peaks comparing pseudoreplicates from replicate 1
- rep2-pr: IDR peaks comparing pseudoreplicates from replicate 2
- repi-repj: IDR peaks comparing true replicates (rep i vs. rep j)
Reproducibility QC and Peak Detection Statistics (Irreproducible Discovery Rate)
| Nt | 0 |
|---|---|
| N1 | 266 |
| N2 | 0 |
| Np | 0 |
| N optimal | 266 |
| N conservative | 266 |
| Optimal Set | self_pseudo_rep_rep1 |
| Conservative Set | self_pseudo_rep_rep1 |
| Rescue Ratio | 0.0 |
| Self Consistency Ratio | 1.0 |
| Reproducibility Test | pass |
- N1: Replicate 1 self-consistent IDR 0.05 peaks (comparing two pseudoreplicates generated by subsampling Rep1 reads)
- N2: Replicate 2 self-consistent IDR 0.05 peaks (comparing two pseudoreplicates generated by subsampling Rep2 reads)
- Nt: True Replicate consistent IDR 0.05 peaks (comparing true replicates Rep1 vs Rep2 )
- Np: Pooled-pseudoreplicate consistent IDR 0.05 peaks (comparing two pseudoreplicates generated by subsampling pooled reads from Rep1 and Rep2 )
- Self-consistency Ratio: max(N1,N2) / min (N1,N2)
- Rescue Ratio: max(Np,Nt) / min (Np,Nt)
- Reproducibility Test: If Self-consistency Ratio >2 AND Rescue Ratio > 2, then 'Fail' else 'Pass'
