Flagstat QC (all)
| Reads (QC-passed) | Reads (QC-failed) | Dupes (QC-passed) | Dupes (QC-failed) | Mapped Reads | % Mapped | |
|---|---|---|---|---|---|---|
| rep1 | 16433956 | 0 | 0 | 0 | 16015681 | 97.45 |
Dup QC (all)
Flagstat QC (all, filtered)
PBC QC (all)
| Total Read Pairs | Distinct Read Pairs | One Read Pair | Two Read Pairs | NRF = Distinct/Total | PBC1 = OnePair/Distinct | PBC2 = OnePair/TwoPair | |
|---|---|---|---|---|---|---|---|
| rep1 | 13749349 | 13250021 | 12823570 | 379681 | 0.963684 | 0.967815 | 33.774590 |
| ctl1 | 97578156 | 96132111 | 94755797 | 1345982 | 0.985181 | 0.985683 | 70.399008 |
NRF (non redundant fraction)
PBC1 (PCR Bottleneck coefficient 1)
PBC2 (PCR Bottleneck coefficient 2)
PBC1 is the primary measure. Provisionally
- 0-0.5 is severe bottlenecking
- 0.5-0.8 is moderate bottlenecking
- 0.8-0.9 is mild bottlenecking
- 0.9-1.0 is no bottlenecking
Cross-correlation QC (all)
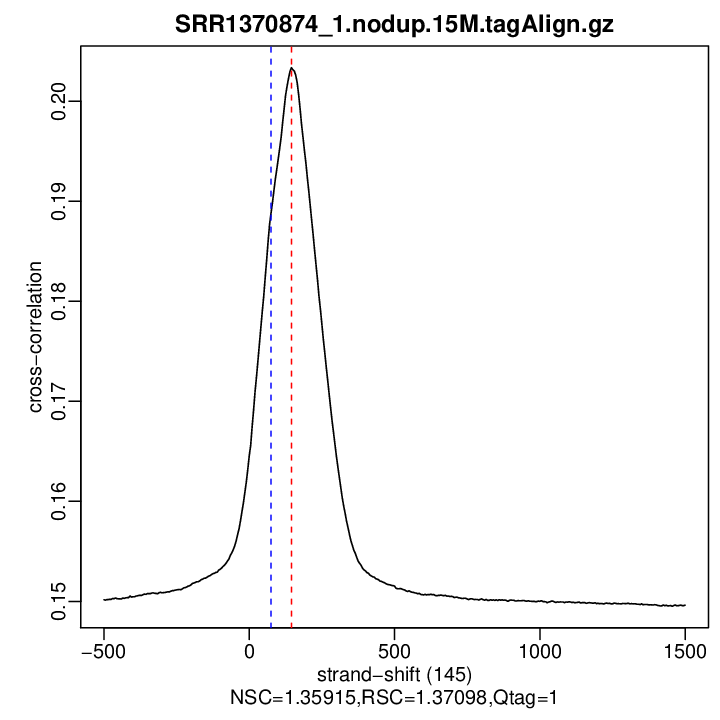
rep1
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | NSC | RSC | |
|---|---|---|---|---|---|---|---|---|---|
| rep1 | 13213324 | 145 | 0.203358157666487 | 75 | 0.1888174 | 1500 | 0.1496215 | 1.359151 | 1.370977 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display
