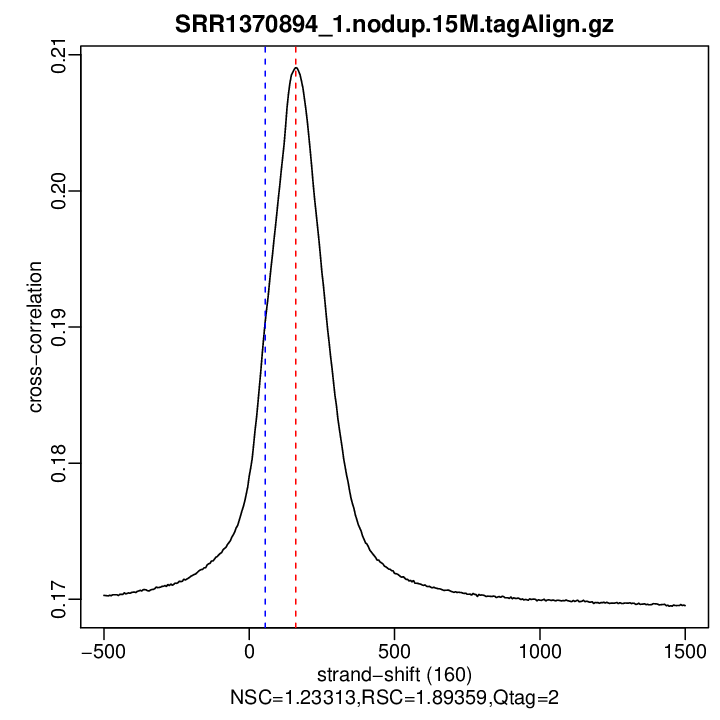
Enrichment QC (strand cross-correlation measures)
| Reads | 15000000 |
|---|---|
| Est. Fragment Len. | 160 |
| Corr. Est. Fragment Len. | 0.209062520474653 |
| Phantom Peak | 55 |
| Corr. Phantom Peak | 0.1904106 |
| Argmin. Corr. | 1500 |
| Min. Corr. | 0.1695375 |
| NSC | 1.233134 |
| RSC | 1.893585 |
NOTE: Reads from replicates are subsampled to a max of 15M
- Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
- Relative strand cross-correlation coefficient (RSC) = col10 in outFile
- Estimated fragment length = col3 in outFile, take the top value