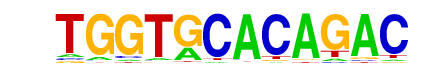
| p-value: | 1e-203 |
| log p-value: | -4.683e+02 |
| Information Content per bp: | 1.703 |
| Number of Target Sequences with motif | 1629.0 |
| Percentage of Target Sequences with motif | 4.02% |
| Number of Background Sequences with motif | 886.0 |
| Percentage of Background Sequences with motif | 1.19% |
| Average Position of motif in Targets | 168.6 +/- 90.6bp |
| Average Position of motif in Background | 216.5 +/- 190.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0026.1_Gm397_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GTCTGTGCACCA---
NNGTATGTGCACATNNN |
|


|
|
PB0104.1_Zscan4_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GTCTGTGCACCA---
NTNTATGTGCACATNNN |
|


|
|
RCS1(MacIsaac)/Yeast
| Match Rank: | 3 |
| Score: | 0.62
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | GTCTGTGCACCA
-----TGCACCC |
|


|
|
PB0130.1_Gm397_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GTCTGTGCACCA
NNGCGTGTGTGCNGCN |
|


|
|
PHYPADRAFT_182268/MA1008.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GTCTGTGCACCA
TTGTCGGTGA---- |
|


|
|
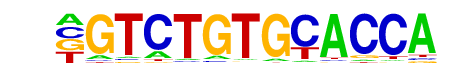
ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GTCTGTGCACCA
WDNCTGGGCA--- |
|

|
|
Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GTCTGTGCACCA
VBSYGTCTGG------ |
|


|
|
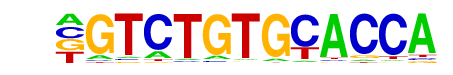
PHYPADRAFT_28324/MA1023.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GTCTGTGCACCA
TGTCGGTG----- |
|

|
|
FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GTCTGTGCACCA
NYYTGTTTACHN |
|


|
|
MET28(MacIsaac)/Yeast
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GTCTGTGCACCA
--CTGTGG---- |
|


|
|