
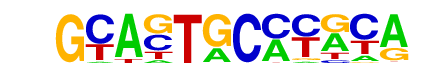













| p-value: | 1e-1231 |
| log p-value: | -2.835e+03 |
| Information Content per bp: | 1.616 |
| Number of Target Sequences with motif | 7584.0 |
| Percentage of Target Sequences with motif | 18.72% |
| Number of Background Sequences with motif | 3479.5 |
| Percentage of Background Sequences with motif | 4.67% |
| Average Position of motif in Targets | 190.8 +/- 104.2bp |
| Average Position of motif in Background | 228.7 +/- 184.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.957 |  | 1e-1102 | -2538.132192 | 17.04% | 4.28% | motif file (matrix) |
| 2 | 0.739 |  | 1e-972 | -2240.156867 | 10.07% | 1.39% | motif file (matrix) |
| 3 | 0.910 |  | 1e-815 | -1878.387280 | 20.99% | 8.12% | motif file (matrix) |
| 4 | 0.673 |  | 1e-416 | -960.000330 | 7.87% | 2.27% | motif file (matrix) |
| 5 | 0.742 |  | 1e-332 | -766.535547 | 13.63% | 6.56% | motif file (matrix) |
| 6 | 0.827 |  | 1e-283 | -653.693746 | 45.08% | 34.24% | motif file (matrix) |
| 7 | 0.688 |  | 1e-209 | -481.923875 | 25.64% | 17.84% | motif file (matrix) |
| 8 | 0.621 |  | 1e-175 | -404.295929 | 27.21% | 19.86% | motif file (matrix) |
| 9 | 0.601 |  | 1e-163 | -376.073698 | 5.15% | 2.11% | motif file (matrix) |
| 10 | 0.731 |  | 1e-98 | -227.395825 | 5.08% | 2.62% | motif file (matrix) |
| 11 | 0.653 |  | 1e-94 | -218.598784 | 3.63% | 1.64% | motif file (matrix) |
| 12 | 0.633 |  | 1e-83 | -193.374460 | 2.83% | 1.20% | motif file (matrix) |
| 13 | 0.612 |  | 1e-53 | -123.509128 | 9.10% | 6.56% | motif file (matrix) |