| p-value: | 1e-138 |
| log p-value: | -3.189e+02 |
| Information Content per bp: | 1.695 |
| Number of Target Sequences with motif | 1814.0 |
| Percentage of Target Sequences with motif | 3.31% |
| Number of Background Sequences with motif | 927.0 |
| Percentage of Background Sequences with motif | 1.26% |
| Average Position of motif in Targets | 171.4 +/- 95.8bp |
| Average Position of motif in Background | 212.2 +/- 214.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
SOK2(MacIsaac)/Yeast
| Match Rank: | 1 |
| Score: | 0.77
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
--TTGCCTGC-- |
|


|
|
SOK2/SOK2_BUT14/4-SUT1(Harbison)/Yeast
| Match Rank: | 2 |
| Score: | 0.72
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
---TNCCTGCA- |
|
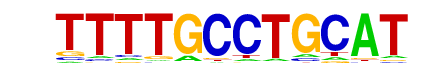

|
|
SOK2/MA0385.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT--
---NNCCTGCAGGT |
|


|
|
AtLEC2(ABI3/VP1)/Arabidopsis thaliana/AthaMap
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
-TTTGCATGGA- |
|
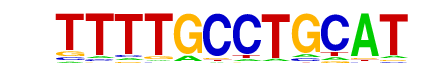

|
|
MOT3/Literature(Harbison)/Yeast
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
---TACCTN--- |
|
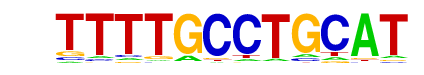

|
|
MOT3(MacIsaac)/Yeast
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
---TACCTN--- |
|
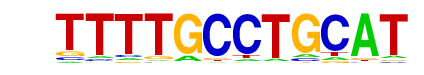

|
|
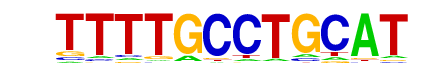
MOT3/MA0340.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
---TACCTN--- |
|

|
|
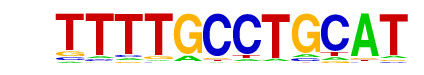
gcm2/MA0917.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
----ACCCGCAT |
|

|
|
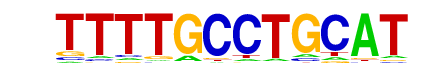
NAC058/MA0938.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.63
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
--TTGCGTGN-- |
|

|
|
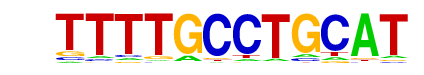
Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo_et_al.)/Homer
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTTTGCCTGCAT
--TTGCGTGCVA |
|

|
|





















