| p-value: | 1e-464 |
| log p-value: | -1.070e+03 |
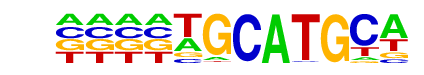
| Information Content per bp: | 1.679 |
| Number of Target Sequences with motif | 3586.0 |
| Percentage of Target Sequences with motif | 6.55% |
| Number of Background Sequences with motif | 1203.9 |
| Percentage of Background Sequences with motif | 1.63% |
| Average Position of motif in Targets | 170.7 +/- 77.9bp |
| Average Position of motif in Background | 214.7 +/- 217.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0091.1_Zbtb3_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.78
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GYACTGCATGCA-
AATCGCACTGCATTCCG |
|


|
|
ABI3/MA0564.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.74
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | GYACTGCATGCA
---CTGCATGCA |
|


|
|
Ddit3::Cebpa/MA0019.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GYACTGCATGCA
GGGATTGCATNN- |
|


|
|
RBFox2(?)/Heart-RBFox2-CLIP-Seq(GSE57926)/Homer
| Match Rank: | 4 |
| Score: | 0.70
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | GYACTGCATGCA
----TGCATGCA |
|

|
|
CTCF-SatelliteElement(Zf?)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | -12
| | Orientation: | reverse strand |
| Alignment: | ------------GYACTGCATGCA
TGGCCANNNNNGGAACTGCA---- |
|


|
|
SOK2/MA0385.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GYACTGCATGCA
-ACCTGCAGGCA |
|


|
|
RCS1/RCS1_H2O2Hi/35-RCS1(Harbison)/Yeast
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GYACTGCATGCA
--ANTGCACCC- |
|


|
|
LEC2/MA0581.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | GYACTGCATGCA---
----TGCATGCACAT |
|


|
|
PHD1/MA0355.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GYACTGCATGCA
-NGNTGCAGGN- |
|


|
|
FUS3/MA0565.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | GYACTGCATGCA-
----CGCATGCGC |
|


|
|





















