| p-value: | 1e-25 |
| log p-value: | -5.935e+01 |
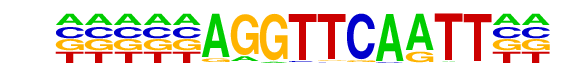
| Information Content per bp: | 1.844 |
| Number of Target Sequences with motif | 1256.0 |
| Percentage of Target Sequences with motif | 2.29% |
| Number of Background Sequences with motif | 1095.4 |
| Percentage of Background Sequences with motif | 1.49% |
| Average Position of motif in Targets | 180.3 +/- 106.1bp |
| Average Position of motif in Background | 238.3 +/- 224.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ct/MA0218.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.73
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AGGTTCAATT
--GTTCAA-- |
|


|
|
Ct/dmmpmm(Noyes_hd)/fly
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGGTTCAATT
NNGTTCAAGN |
|


|
|
ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTTCAATT--
AGTTTCAGTTTC |
|


|
|
PB0030.1_Hnf4a_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----AGGTTCAATT--
CTCCAGGGGTCAATTGA |
|

|
|
STAT1::STAT2/MA0517.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGGTTCAATT---
TCAGTTTCATTTTCC |
|


|
|
TEIL(AP2/EREBP)/Nicotiana tabacum/AthaMap
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGGTTCAATT
AGGTACAT-- |
|


|
|
YHP1/MA0426.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | AGGTTCAATT-
-----CAATTA |
|


|
|
YHP1/Literature(Harbison)/Yeast
| Match Rank: | 8 |
| Score: | 0.64
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | AGGTTCAATT-
-----CAATTA |
|


|
|
YHP1(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.64
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | AGGTTCAATT-
-----CAATTA |
|


|
|
PU.1:IRF8(ETS:IRF)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer
| Match Rank: | 10 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGGTTCAATT--
ASTTTCACTTCC |
|


|
|





















