| p-value: | 1e-68 |
| log p-value: | -1.580e+02 |
| Information Content per bp: | 1.568 |
| Number of Target Sequences with motif | 1137.0 |
| Percentage of Target Sequences with motif | 2.08% |
| Number of Background Sequences with motif | 665.2 |
| Percentage of Background Sequences with motif | 0.90% |
| Average Position of motif in Targets | 174.2 +/- 110.7bp |
| Average Position of motif in Background | 217.9 +/- 211.1bp |
| Strand Bias (log2 ratio + to - strand density) | 0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ACE2/ACE2_YPD/2-SWI5(Harbison)/Yeast
| Match Rank: | 1 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | forward strand |
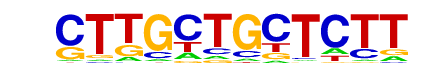
| Alignment: | CTTGCTGCTCTT
--TGCTGGT--- |
|


|
|
Aef1/dmmpmm(Bergman)/fly
| Match Rank: | 2 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTTGCTGCTCTT
-TTGTTG----- |
|


|
|
GLI3(Zf)/Limb-GLI3-ChIP-Chip(GSE11077)/Homer
| Match Rank: | 3 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTTGCTGCTCTT
CGTGGGTGGTCC- |
|


|
|
SWI5/MA0402.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CTTGCTGCTCTT
--TGCTGGTT-- |
|

|
|
MAC1(MacIsaac)/Yeast
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTTGCTGCTCTT
-TTTTCGCTCA- |
|


|
|
grh/dmmpmm(Papatsenko)/fly
| Match Rank: | 6 |
| Score: | 0.56
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTTGCTGCTCTT
-TACCTGCT--- |
|


|
|
Eip74EF/dmmpmm(Pollard)/fly
| Match Rank: | 7 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTTGCTGCTCTT
ACTTCCTGTT--- |
|


|
|
SWI5(MacIsaac)/Yeast
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CTTGCTGCTCTT
--TGCTGGTT-- |
|


|
|
odd/MA0454.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTTGCTGCTCTT
-CTGCTACTGTT |
|


|
|
ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTTGCTGCTCTT
ACTTCCTBGT--- |
|


|
|





















