| p-value: | 1e-129 |
| log p-value: | -2.971e+02 |
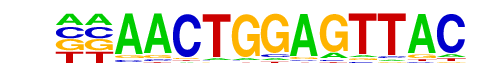
| Information Content per bp: | 1.690 |
| Number of Target Sequences with motif | 1915.0 |
| Percentage of Target Sequences with motif | 4.21% |
| Number of Background Sequences with motif | 1357.2 |
| Percentage of Background Sequences with motif | 1.82% |
| Average Position of motif in Targets | 172.4 +/- 91.4bp |
| Average Position of motif in Background | 233.9 +/- 205.6bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
kni/dmmpmm(SeSiMCMC)/fly
| Match Rank: | 1 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AACTGGAGTTAC
AAAACTGGAACAA- |
|


|
|
YLR278C/MA0430.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.59
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AACTGGAGTTAC
--CCGGAGTT-- |
|


|
|
ARG81/MA0272.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | AACTGGAGTTAC
----NGAGTCAC |
|


|
|
ARG81(MacIsaac)/Yeast
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | AACTGGAGTTAC
----NGAGTCAC |
|


|
|
URC2/MA0422.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AACTGGAGTTAC
--CCGGAGATAA |
|


|
|
LXRE(NR),DR4/RAW-LXRb.biotin-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AACTGGAGTTAC--
TGACCTNTAGTAACCC |
|


|
|
prd/dmmpmm(Down)/fly
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AACTGGAGTTAC
AAACTG------- |
|


|
|
kni/dmmpmm(Bigfoot)/fly
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AACTGGAGTTAC
AAAANTGGANCAA- |
|

|
|
ETV2/MA0762.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AACTGGAGTTAC
AACCGGAAATA- |
|


|
|
HAP1/MA0312.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | AACTGGAGTTAC
---CGGAGATA- |
|


|
|





















