
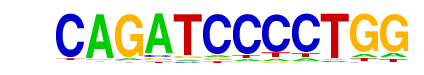









| p-value: | 1e-171 |
| log p-value: | -3.958e+02 |
| Information Content per bp: | 1.746 |
| Number of Target Sequences with motif | 14013.0 |
| Percentage of Target Sequences with motif | 30.77% |
| Number of Background Sequences with motif | 17487.9 |
| Percentage of Background Sequences with motif | 23.41% |
| Average Position of motif in Targets | 186.8 +/- 112.8bp |
| Average Position of motif in Background | 225.8 +/- 189.1bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.18 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.685 |  | 1e-166 | -382.504782 | 3.26% | 1.01% | motif file (matrix) |
| 2 | 0.711 |  | 1e-113 | -262.001861 | 2.98% | 1.13% | motif file (matrix) |
| 3 | 0.688 |  | 1e-100 | -231.365471 | 3.36% | 1.47% | motif file (matrix) |
| 4 | 0.600 |  | 1e-75 | -174.235813 | 9.22% | 6.31% | motif file (matrix) |
| 5 | 0.724 |  | 1e-63 | -146.392404 | 3.16% | 1.65% | motif file (matrix) |
| 6 | 0.710 |  | 1e-61 | -142.543155 | 5.92% | 3.82% | motif file (matrix) |
| 7 | 0.644 |  | 1e-55 | -126.751746 | 3.55% | 2.04% | motif file (matrix) |
| 8 | 0.794 |  | 1e-30 | -70.849975 | 9.93% | 7.97% | motif file (matrix) |
| 9 | 0.701 |  | 1e-21 | -48.586241 | 1.59% | 0.96% | motif file (matrix) |