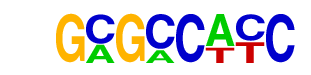
| p-value: | 1e-251 |
| log p-value: | -5.796e+02 |
| Information Content per bp: | 1.829 |
| Number of Target Sequences with motif | 9690.0 |
| Percentage of Target Sequences with motif | 21.28% |
| Number of Background Sequences with motif | 10231.7 |
| Percentage of Background Sequences with motif | 13.70% |
| Average Position of motif in Targets | 190.8 +/- 116.6bp |
| Average Position of motif in Background | 217.8 +/- 173.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.06 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
CTCF/MA0531.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.84
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------GRTGGCGC-
CCGCTAGATGGCGCC |
|


|
|
RPN4/MA0373.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GRTGGCGC
GGTGGCG- |
|


|
|
RAP2-10/MA0980.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.75
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GRTGGCGC
GGCGGCGC |
|


|
|
brk/dmmpmm(Pollard)/fly
| Match Rank: | 4 |
| Score: | 0.75
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GRTGGCGC-
-CTGGCGCC |
|


|
|
brk/MA0213.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.75
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GRTGGCGC-
-CTGGCGCC |
|


|
|
brk/dmmpmm(Papatsenko)/fly
| Match Rank: | 6 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GRTGGCGC-
-CTGGCGCC |
|


|
|
RAP2-6/MA1052.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.74
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GRTGGCGC
GGCGGCGC |
|


|
|
ERF109/MA1053.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.73
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GRTGGCGC
GGCGGCGC |
|


|
|
brk/dmmpmm(Bigfoot)/fly
| Match Rank: | 9 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GRTGGCGC-
--CGGCGCC |
|


|
|
CRF4/MA0976.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.72
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GRTGGCGC
GGCGGCGC |
|


|
|