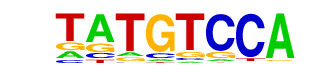
| p-value: | 1e-5 |
| log p-value: | -1.322e+01 |
| Information Content per bp: | 1.614 |
| Number of Target Sequences with motif | 6150.0 |
| Percentage of Target Sequences with motif | 13.51% |
| Number of Background Sequences with motif | 9395.9 |
| Percentage of Background Sequences with motif | 12.58% |
| Average Position of motif in Targets | 188.8 +/- 116.5bp |
| Average Position of motif in Background | 230.1 +/- 206.6bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0104.1_Zscan4_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TGGACATA---
TACATGTGCACATAAAA |
|


|
|
PB0026.1_Gm397_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TGGACATA---
CAGATGTGCACATACGT |
|


|
|
PB0134.1_Hnf4a_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGGACATA----
NNATTGGACTTTNGNN |
|


|
|
ZNF354C/MA0130.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGGACATA
GTGGAT--- |
|


|
|
NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer
| Match Rank: | 5 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGGACATA
AATGGAAAAT |
|


|
|
PB0133.1_Hic1_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.66
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TGGACATA---
NNNNTTGGGCACNNCN |
|


|
|
NFATC2/MA0152.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGGACATA
TGGAAAA- |
|


|
|
NFATC1/MA0624.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGGACATA
NNTGGAAANN |
|


|
|
NFATC3/MA0625.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGGACATA
AATGGAAAAT |
|


|
|
HIC2/MA0738.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGGACATA
NGTGGGCAT- |
|


|
|