Flagstat QC (all)
Dup QC (all)
Flagstat QC (all, filtered)
PBC QC (all)
| Total Read Pairs | Distinct Read Pairs | One Read Pair | Two Read Pairs | NRF = Distinct/Total | PBC1 = OnePair/Distinct | PBC2 = OnePair/TwoPair | |
|---|---|---|---|---|---|---|---|
| rep1 | 23168913 | 20740263 | 18569839 | 1953352 | 0.895176 | 0.895352 | 9.506653 |
| ctl1 | 16340490 | 7720329 | 4661764 | 1212451 | 0.472466 | 0.603830 | 3.844909 |
NRF (non redundant fraction)
PBC1 (PCR Bottleneck coefficient 1)
PBC2 (PCR Bottleneck coefficient 2)
PBC1 is the primary measure. Provisionally
- 0-0.5 is severe bottlenecking
- 0.5-0.8 is moderate bottlenecking
- 0.8-0.9 is mild bottlenecking
- 0.9-1.0 is no bottlenecking
Cross-correlation QC (all)
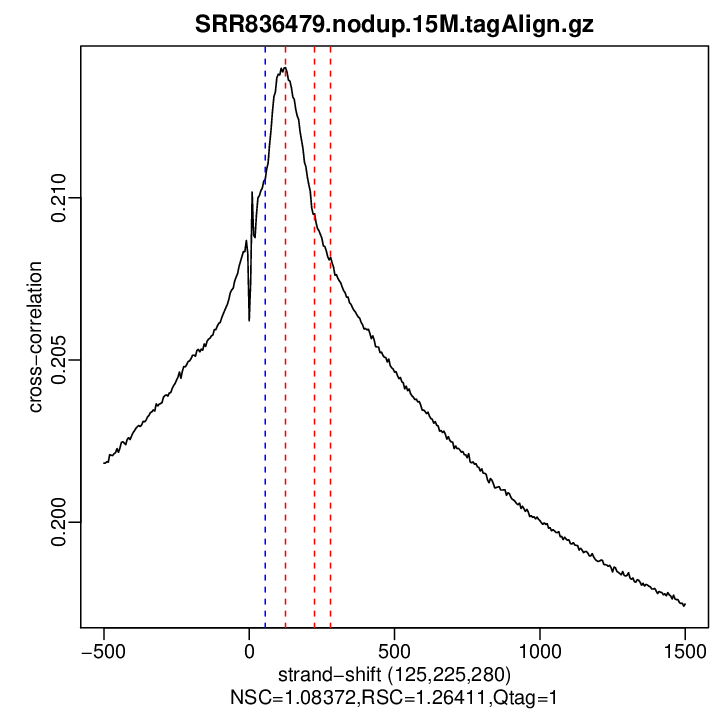
rep1
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | NSC | RSC | |
|---|---|---|---|---|---|---|---|---|---|
| rep1 | 15000000 | 125 | 0.214013912111025 | 55 | 0.2105595 | 1500 | 0.1974803 | 1.083723 | 1.264113 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display

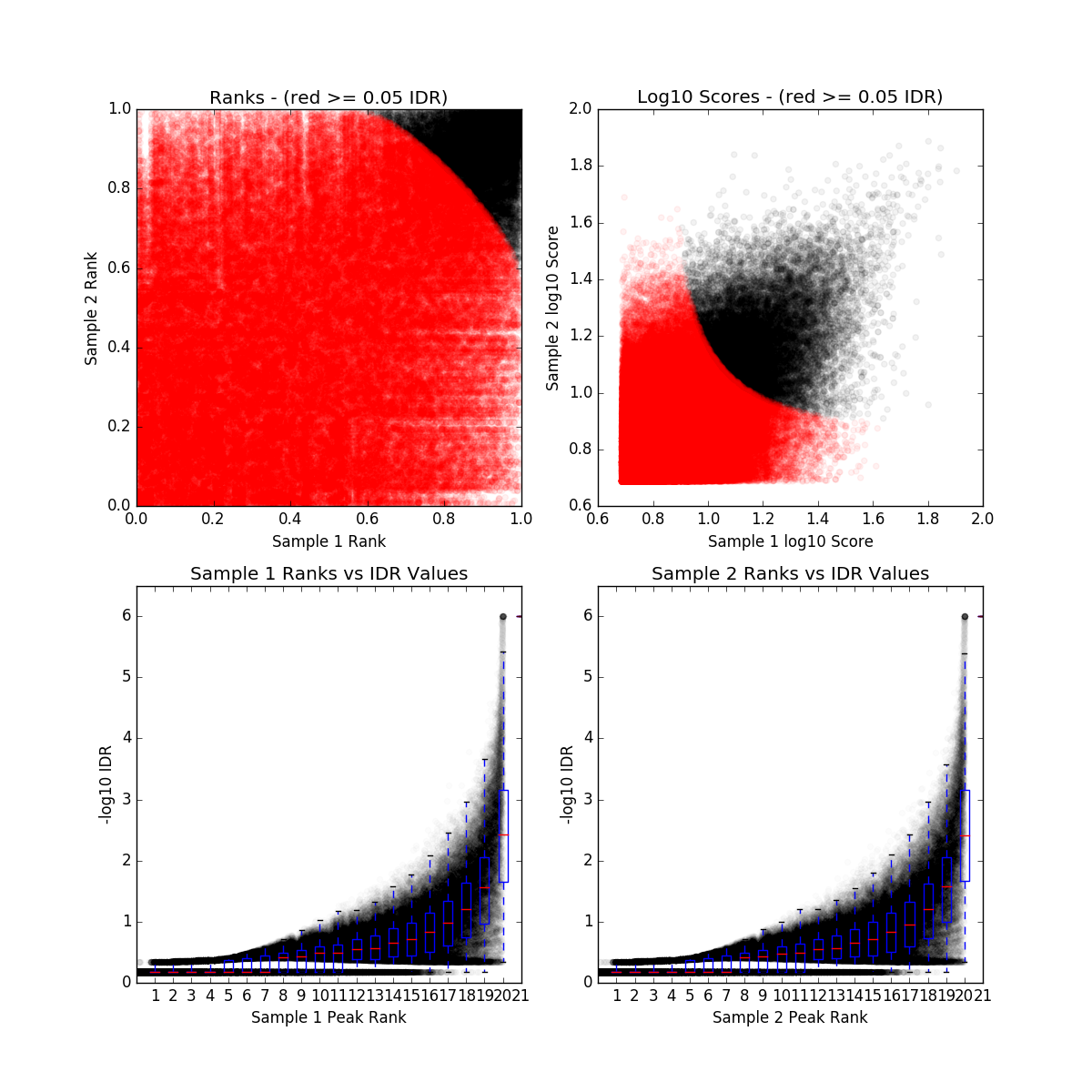
IDR QC (idr)
SRR836479_IDR_final.qc
| Nt | N1 | Np | conservative_set | optimal_set | rescue_ratio | self_consistency_ratio | reproducibility |
|---|---|---|---|---|---|---|---|
| 0 | 17815 | 0 | N/A | N/A | NaN | 1.0 | 1 |